Diversity, functional classification and genotyping of SHV β-lactamases in Klebsiella pneumoniae
- PMID: 39432416
- PMCID: PMC11493186
- DOI: 10.1099/mgen.0.001294
Diversity, functional classification and genotyping of SHV β-lactamases in Klebsiella pneumoniae
Abstract
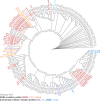
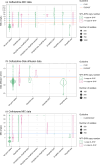
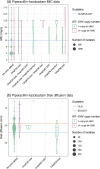
Interpreting the phenotypes of bla SHV alleles in Klebsiella pneumoniae genomes is complex. Whilst all strains are expected to carry a chromosomal copy conferring resistance to ampicillin, they may also carry mutations in chromosomal bla SHV alleles or additional plasmid-borne bla SHV alleles that have extended-spectrum β-lactamase (ESBL) activity and/or β-lactamase inhibitor (BLI) resistance activity. In addition, the role of individual mutations/a changes is not completely documented or understood. This has led to confusion in the literature and in antimicrobial resistance (AMR) gene databases [e.g. the National Center for Biotechnology Information (NCBI) Reference Gene Catalog and the β-lactamase database (BLDB)] over the specific functionality of individual sulfhydryl variable (SHV) protein variants. Therefore, the identification of ESBL-producing strains from K. pneumoniae genome data is complicated. Here, we reviewed the experimental evidence for the expansion of SHV enzyme function associated with specific aa substitutions. We then systematically assigned SHV alleles to functional classes (WT, ESBL and BLI resistant) based on the presence of these mutations. This resulted in the re-classification of 37 SHV alleles compared with the current assignments in the NCBI's Reference Gene Catalog and/or BLDB (21 to WT, 12 to ESBL and 4 to BLI resistant). Phylogenetic and comparative genomic analyses support that (i) SHV-1 (encoded by bla SHV-1) is the ancestral chromosomal variant, (ii) ESBL- and BLI-resistant variants have evolved multiple times through parallel substitution mutations, (iii) ESBL variants are mostly mobilized to plasmids and (iv) BLI-resistant variants mostly result from mutations in chromosomal bla SHV. We used matched genome-phenotype data from the KlebNET-GSP AMR Genotype-Phenotype Group to identify 3999 K. pneumoniae isolates carrying one or more bla SHV alleles but no other acquired β-lactamases to assess genotype-phenotype relationships for bla SHV. This collection includes human, animal and environmental isolates collected between 2001 and 2021 from 24 countries. Our analysis supports that mutations at Ambler sites 238 and 179 confer ESBL activity, whilst most omega-loop substitutions do not. Our data also provide support for the WT assignment of 67 protein variants, including 8 that were noted in public databases as ESBL. These eight variants were reclassified as WT because they lack ESBL-associated mutations, and our phenotype data support susceptibility to third-generation cephalosporins (SHV-27, SHV-38, SHV-40, SHV-41, SHV-42, SHV-65, SHV-164 and SHV-187). The approach and results outlined here have been implemented in Kleborate v2.4.1 (a software tool for genotyping K. pneumoniae), whereby known and novel bla SHV alleles are classified based on causative mutations. Kleborate v2.4.1 was updated to include ten novel protein variants from the KlebNET-GSP dataset and all alleles in public databases as of November 2023. This study demonstrates the power of sharing AMR phenotypes alongside genome data to improve the understanding of resistance mechanisms.
Keywords: AMR; BLI resistance; K. pneumoniae; SHV; extended-spectrum β-lactamase (ESBL); genotype; prediction; β-lactamase.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures




References
-
- Hammond DS, Schooneveldt JM, Nimmo GR, Huygens F, Giffard PM bla. bla(SHV) genes in Klebsiella pneumoniae: different allele distributions are associated with different promoters within individual isolates. Antimicrob Agents Chemother. 2005;49:256–263. doi: 10.1128/AAC.49.1.256-263.2005. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

