CD4 T cells restricted to DRB1*15:01 recognize two Epstein-Barr virus glycoproteins capable of intracellular antigen presentation
- PMID: 39432795
- PMCID: PMC11536159
- DOI: 10.1073/pnas.2416097121
CD4 T cells restricted to DRB1*15:01 recognize two Epstein-Barr virus glycoproteins capable of intracellular antigen presentation
Abstract
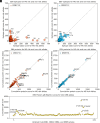
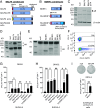
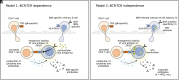
Both genetic and environmental factors contribute to multiple sclerosis (MS) risk. Infection with the Epstein-Barr virus (EBV) is the strongest environmental risk factor, and HLA-DR15 is the strongest genetic risk factor for MS. We employed computational methods and in vitro assays for CD4 T cell activation to investigate the DR15-restricted response to EBV. Using a machine learning-based HLA ligand predictor, the EBV glycoprotein B (gB) was predicted to be enriched in epitopes restricted to presentation by DRB1*15:01. In DR15-positive individuals, two epitopes comprised the major CD4 T cell response to gB. Surprisingly, the expression of recombinant gB in a DR15-homozygous B cell line or primary autologous B cells elicited a CD4 T cell response, indicating that intracellular gB was loaded onto HLA class II molecules. By deleting the signal sequence of gB, we determined that this pathway for direct activation of CD4 T cells was dependent on trafficking to the endoplasmic reticulum (ER) within the B cell. We screened seven recombinant EBV antigens from the ER compartment for immune responses in DR15-negative vs. DR15-homozygous individuals. In addition to gB, gH was a key CD4 T cell target in individuals homozygous for DR15. Compared to non-DR15 controls, DR15-homozygotes had significantly higher T cell responses to both gB and gH but not to EBV latent or lytic antigens overall. Responses to gB and gH were slightly elevated in DR15 homozygotes with MS. Our results link MS environmental and genetic risk factors by demonstrating that HLA-DR15 dictates CD4 T cell immunity to EBV antigens.
Keywords: Epstein–Barr virus; HLA-DR15; antivirals; multiple sclerosis.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures


Comment in
-
Connecting the dots: Presentation of EBV antigens on HLA class II risk alleles connects the two main risk factors of multiple sclerosis.Proc Natl Acad Sci U S A. 2024 Dec 3;121(49):e2420070121. doi: 10.1073/pnas.2420070121. Epub 2024 Nov 25. Proc Natl Acad Sci U S A. 2024. PMID: 39585999 Free PMC article. No abstract available.
References
-
- Bjornevik K., et al. , Longitudinal analysis reveals high prevalence of Epstein–Barr virus associated with multiple sclerosis. Science 375, 296–301 (2022). - PubMed
-
- Vietzen H., et al. , Ineffective control of Epstein–Barr-virus-induced autoimmunity increases the risk for multiple sclerosis. Cell 186, 5705–5718.e13 (2023). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

