Nucleoprotein Phase-Separation Affinities Revealed via Atomistic Simulations of Short Peptide and RNA Fragments
- PMID: 39432826
- PMCID: PMC11972660
- DOI: 10.1021/acs.jpclett.4c02654
Nucleoprotein Phase-Separation Affinities Revealed via Atomistic Simulations of Short Peptide and RNA Fragments
Abstract
Liquid-liquid phase separation of proteins and nucleic acids into condensate phases is a versatile mechanism for ensuring the compartmentalization of cellular biochemistry. RNA molecules play critical roles in these condensates, particularly in transcriptional regulation and stress responses, exhibiting a wide range of thermodynamic and dynamic behaviors. However, deciphering the molecular grammar that governs the stability and dynamics of protein-RNA condensates remains challenging due to the multicomponent and heterogeneous nature of condensates. In this study, we employ atomistic simulations of 20 distinct mixtures containing minimal RNA and peptide fragments which allows us to dissect the phase-separating affinities of all 20 amino acids in the presence of RNA. Our findings elucidate chemically specific interactions, hydration profiles, and ionic effects that synergistically promote or suppress protein-RNA phase separation. We map a ternary phase diagram of interactions, identifying four distinct groups of residues that promote, maintain, suppress, and disrupt protein-RNA clusters.
Figures


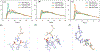
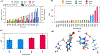
Update of
-
Nucleoprotein phase-separation affinities revealed via atomistic simulations of short peptide and RNA fragments.bioRxiv [Preprint]. 2024 Sep 26:2024.09.24.614800. doi: 10.1101/2024.09.24.614800. bioRxiv. 2024. Update in: J Phys Chem Lett. 2024 Oct 31;15(43):10811-10817. doi: 10.1021/acs.jpclett.4c02654. PMID: 39386696 Free PMC article. Updated. Preprint.
Similar articles
-
Nucleoprotein phase-separation affinities revealed via atomistic simulations of short peptide and RNA fragments.bioRxiv [Preprint]. 2024 Sep 26:2024.09.24.614800. doi: 10.1101/2024.09.24.614800. bioRxiv. 2024. Update in: J Phys Chem Lett. 2024 Oct 31;15(43):10811-10817. doi: 10.1021/acs.jpclett.4c02654. PMID: 39386696 Free PMC article. Updated. Preprint.
-
Surfactants or scaffolds? RNAs of varying lengths control the thermodynamic stability of condensates differently.Biophys J. 2023 Jul 25;122(14):2973-2987. doi: 10.1016/j.bpj.2023.03.006. Epub 2023 Mar 6. Biophys J. 2023. PMID: 36883003 Free PMC article.
-
Submillisecond Atomistic Molecular Dynamics Simulations Reveal Hydrogen Bond-Driven Diffusion of a Guest Peptide in Protein-RNA Condensate.J Phys Chem B. 2024 Mar 14;128(10):2347-2359. doi: 10.1021/acs.jpcb.3c08126. Epub 2024 Feb 28. J Phys Chem B. 2024. PMID: 38416758 Free PMC article.
-
Using quantitative reconstitution to investigate multicomponent condensates.RNA. 2022 Jan;28(1):27-35. doi: 10.1261/rna.079008.121. Epub 2021 Nov 12. RNA. 2022. PMID: 34772789 Free PMC article. Review.
-
Rich Phase Separation Behavior of Biomolecules.Mol Cells. 2022 Jan 31;45(1):6-15. doi: 10.14348/molcells.2021.0204. Mol Cells. 2022. PMID: 34966005 Free PMC article. Review.
References
-
- Hirose T, Ninomiya K, Nakagawa S, Yamazaki T. A guide to membraneless organelles and their various roles in gene regulation. Nat Rev Mol Cell Biol. 2023;24: 288–304. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

