Breast cancer secretes anti-ferroptotic MUFAs and depends on selenoprotein synthesis for metastasis
- PMID: 39433871
- PMCID: PMC11555046
- DOI: 10.1038/s44321-024-00142-x
Breast cancer secretes anti-ferroptotic MUFAs and depends on selenoprotein synthesis for metastasis
Abstract
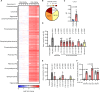
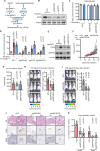
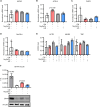
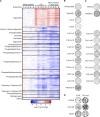
The limited availability of therapeutic options for patients with triple-negative breast cancer (TNBC) contributes to the high rate of metastatic recurrence and poor prognosis. Ferroptosis is a type of cell death caused by iron-dependent lipid peroxidation and counteracted by the antioxidant activity of the selenoprotein GPX4. Here, we show that TNBC cells secrete an anti-ferroptotic factor in the extracellular environment when cultured at high cell densities but are primed to ferroptosis when forming colonies at low density. We found that secretion of the anti-ferroptotic factors, identified as monounsaturated fatty acid (MUFA) containing lipids, and the vulnerability to ferroptosis of single cells depends on the low expression of stearyl-CoA desaturase (SCD) that is proportional to cell density. Finally, we show that the inhibition of Sec-tRNAsec biosynthesis, an essential step for selenoprotein production, causes ferroptosis and impairs the lung seeding of circulating TNBC cells that are no longer protected by the MUFA-rich environment of the primary tumour.
Keywords: Breast Cancer; Ferroptosis; Lipid Metabolism; Metastasis; Selenium Metabolism.
© 2024. The Author(s).
Conflict of interest statement
Figures







References
-
- Burk RF, Hill KE (2015) Regulation of Selenium Metabolism and Transport. Annu Rev Nutr 35:109–134 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

