Comprehensive multi-omics integration uncovers mitochondrial gene signatures for prognosis and personalized therapy in lung adenocarcinoma
- PMID: 39434164
- PMCID: PMC11492473
- DOI: 10.1186/s12967-024-05754-y
Comprehensive multi-omics integration uncovers mitochondrial gene signatures for prognosis and personalized therapy in lung adenocarcinoma
Abstract
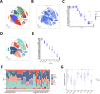
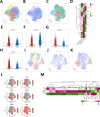
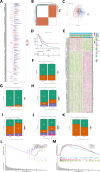
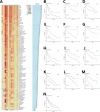
The therapeutic efficacy of lung adenocarcinoma (LUAD), the most prevalent histological subtype of primary lung cancer, remains inadequate, with accurate prognostic assessment posing significant challenges. This study sought to elucidate the prognostic significance of mitochondrial-related genes in LUAD through an integrative multi-omics approach, aimed at developing personalized therapeutic strategies. Utilizing transcriptomic and single-cell RNA sequencing (scRNA-seq) data, alongside clinical information from publicly available databases, we first applied dimensionality reduction and clustering techniques to the LUAD single-cell dataset, focusing on the subclassification of fibroblasts, epithelial cells, and T cells. Mitochondrial-related prognostic genes were subsequently identified using TCGA-LUAD data, and LUAD cases were stratified into distinct molecular subtypes through consensus clustering, allowing for the exploration of gene expression profiles and clinical feature distributions across subtypes. By leveraging an ensemble of machine learning algorithms, we developed an Artificial Intelligence-Derived Prognostic Signature (AIDPS) model based on mitochondrial-related genes and validated its prognostic accuracy across multiple independent datasets. The AIDPS model demonstrated robust predictive power for LUAD patient outcomes, revealing significant differences in responses to immunotherapy and chemotherapy, as well as survival outcomes between risk groups. Furthermore, we conducted comprehensive analyses of tumor mutation burden (TMB), immune microenvironment characteristics, and genome-wide association study (GWAS) data, providing additional insights into the mechanistic roles of mitochondrial-related genes in LUAD pathogenesis. This study not only offers a novel approach to improving prognostic assessments in LUAD but also establishes a strong foundation for the development of personalized therapeutic interventions.
Keywords: Lung adenocarcinoma; Machine learning; Mitochondrial-related genes; Prognostic model; Single nucleotide variation.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no conflicts of interest to report regarding the present study.
Figures




References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer statistics 2020: GLOBOCAN estimates of incidence and Mortality Worldwide for 36 cancers in 185 countries. Cancer J Clin. 2021;71(3):209–49. - PubMed
-
- Xu Y, Krishnan A, Wan XS, Majima H, Yeh CC, Ludewig G, et al. Mutations in the promoter reveal a cause for the reduced expression of the human manganese superoxide dismutase gene in cancer cells. Oncogene. 1999;18(1):93–102. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

