Host-microbe interaction-mediated resistance to DSS-induced inflammatory enteritis in sheep
- PMID: 39434180
- PMCID: PMC11492479
- DOI: 10.1186/s40168-024-01932-8
Host-microbe interaction-mediated resistance to DSS-induced inflammatory enteritis in sheep
Abstract
Background: The disease resistance phenotype is closely related to immunomodulatory function and immune tolerance and has far-reaching implications in animal husbandry and human health. Microbes play an important role in the initiation, prevention, and treatment of diseases, but the mechanisms of host-microbiota interactions in disease-resistant phenotypes are poorly understood. In this study, we hope to uncover and explain the role of microbes in intestinal diseases and their mechanisms of action to identify new potential treatments.
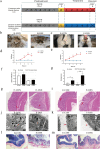
Methods: First, we established the colitis model of DSS in two breeds of sheep and then collected the samples for multi-omics testing including metagenes, metabolome, and transcriptome. Next, we made the fecal bacteria liquid from the four groups of sheep feces collected from H-CON, H-DSS, E-CON, and E-DSS to transplant the fecal bacteria into mice. H-CON feces were transplanted into mice named HH group and H-DSS feces were transplanted into mice named HD group and Roseburia bacteria treatment named HDR groups. E-CON feces were transplanted into mice named EH group and E-DSS feces were transplanted into mice in the ED group and Roseburia bacteria treatment named EDR groups. After successful modeling, samples were taken for multi-omics testing. Finally, colitis mice in HD group and ED group were administrated with Roseburia bacteria, and the treatment effect was evaluated by H&E, PAS, immunohistochemistry, and other experimental methods.
Results: The difference in disease resistance of sheep to DSS-induced colitis disease is mainly due to the increase in the abundance of Roseburia bacteria and the increase of bile acid secretion in the intestinal tract of Hu sheep in addition to the accumulation of potentially harmful bacteria in the intestine when the disease occurs, which makes the disease resistance of Hu sheep stronger under the same disease conditions. However, the enrichment of harmful microorganisms in East Friesian sheep activated the TNFα signalling pathway, which aggravated the intestinal injury, and then the treatment of FMT mice by culturing Roseburia bacteria found that Roseburia bacteria had a good curative effect on colitis.
Conclusion: Our study showed that in H-DSS-treated sheep, the intestinal barrier is stabilized with an increase in the abundance of beneficial microorganisms. Our data also suggest that Roseburia bacteria have a protective effect on the intestinal barrier of Hu sheep. Accumulating evidence suggests that host-microbiota interactions are associated with IBD disease progression. Video Abstract.
Keywords: Roseburia bacteria; Fecal microbiota transplantation (FMT); Gut microbiota; Intestinal inflammation.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Chen L, et al. The long-term genetic stability and individual specificity of the human gut microbiome. Cell. 2021;184:2302-2315.e2312. 10.1016/j.cell.2021.03.024. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

