The STAT3/SETDB2 axis dictates NF-κB-mediated inflammation in macrophages during wound repair
- PMID: 39435663
- PMCID: PMC11530128
- DOI: 10.1172/jci.insight.179017
The STAT3/SETDB2 axis dictates NF-κB-mediated inflammation in macrophages during wound repair
Abstract
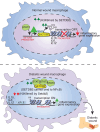
Macrophage transition from an inflammatory to reparative phenotype after tissue injury is controlled by epigenetic enzymes that regulate inflammatory gene expression. We have previously identified that the histone methyltransferase SETDB2 in macrophages drives tissue repair by repressing NF-κB-mediated inflammation. Complementary ATAC-Seq and RNA-Seq of wound macrophages isolated from mice deficient in SETDB2 in myeloid cells revealed that SETDB2 suppresses the inflammatory gene program by inhibiting chromatin accessibility at NF-κB-dependent gene promoters. We found that STAT3 was required for SETDB2 expression in macrophages, yet paradoxically, it also functioned as a binding partner of SETDB2 where it repressed SETDB2 activity by inhibiting its interaction with the NF-κB component, RELA, leading to increased RELA/NF-κB-mediated inflammatory gene expression. Furthermore, RNA-Seq in wound macrophages from STAT3-deficient mice corroborated this and revealed STAT3 and SETDB2 transcriptionally coregulate overlapping genes. Finally, in diabetic wound macrophages, STAT3 expression and STAT3/SETDB2 binding were increased. We have identified what we believe to be a novel STAT3/SETDB2 axis that modulates macrophage phenotype during tissue repair and may be an important therapeutic target for nonhealing diabetic wounds.
Keywords: Inflammation; Transcription.
Conflict of interest statement
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

