Circulating proteins linked to apoptosis processes and fast development of end-stage kidney disease in diabetes
- PMID: 39435665
- PMCID: PMC11529980
- DOI: 10.1172/jci.insight.178373
Circulating proteins linked to apoptosis processes and fast development of end-stage kidney disease in diabetes
Abstract
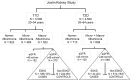
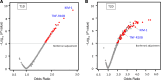
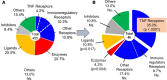
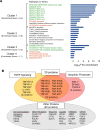
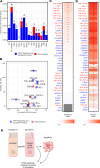
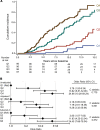
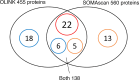
Many circulating proteins are associated with risk of ESKD, but their source and the biological pathways/disease processes they represent are unclear. Using OLINK proteomics platform, concentrations of 455 proteins were measured in plasma specimens obtained at baseline from 399 individuals with diabetes. Elevated concentrations of 46 circulating proteins were associated (P < 1 × 10-5) with development of ESKD (n = 143) during 7-15 years of follow-up. Twenty of these proteins enriched apoptosis/TNF receptor signaling pathways. A subset of 20 proteins (5-7 proteins), summarized as an apoptosis score, together with clinical variables accurately predicted risk of ESKD. Expression of genes encoding the 46 proteins in peripheral WBCs showed no difference between cells from individuals who did or did not develop ESKD. In contrast, plasma concentration of many of the 46 proteins differed by this outcome. In single-nucleus RNA-Seq analysis of kidney biopsies, the majority of genes encoding for the 20 apoptosis/TNF receptor proteins were overexpressed in injured versus healthy proximal tubule cells. Expression of these 20 genes also correlated with the overall index of apoptosis in these cells. Elevated levels of circulating proteins flagging apoptotic processes/TNF receptor signaling pathways - and likely originating from kidney cells, including injured/apoptotic proximal tubular cells - preceded the development of ESKD.
Keywords: Apoptosis; Diabetes; Nephrology.
Conflict of interest statement
Figures


References
-
- NIH. United States Renal Data System. https://www.niddk.nih.gov/about-niddk/strategic-plans-reports/usrds Accessed September 4, 2024.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

