Intra-host genomic diversity and integration landscape of human tissue-resident DNA virome
- PMID: 39436041
- PMCID: PMC11602146
- DOI: 10.1093/nar/gkae871
Intra-host genomic diversity and integration landscape of human tissue-resident DNA virome
Abstract
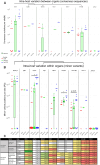
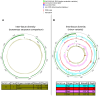
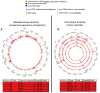
The viral intra-host genetic diversities and interactions with the human genome during decades of persistence remain poorly characterized. In this study, we analyzed the variability and integration sites of persisting viruses in nine organs from thirteen individuals who died suddenly from non-viral causes. The viruses studied included parvovirus B19, six herpesviruses, Merkel cell (MCPyV) and JC polyomaviruses, totaling 127 genomes. The viral sequences across organs were remarkably conserved within each individual, suggesting that persistence stems from single dominant strains. This indicates that intra-host viral evolution, thus far inferred primarily from immunocompromised patients, is likely overestimated in healthy subjects. Indeed, we detected increased viral subpopulations in two individuals with putative reactivations, suggesting that replication status influences diversity. Furthermore, we identified asymmetrical mutation patterns reflecting selective pressures exerted by the host. Strikingly, our analysis revealed non-clonal viral integrations even in individuals without cancer. These included MCPyV integrations and truncations resembling clonally expanded variants in Merkel cell carcinomas, as well as novel junctions between herpesvirus 6B and mitochondrial sequences, the significance of which remains to be evaluated. Our work systematically characterizes the genomic landscape of the tissue-resident virome, highlighting potential deviations occurring during disease.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





References
-
- Thomson E., Ip C.L.C., Badhan A., Christiansen M.T., Adamson W., Ansari M.A., Bibby D., Breuer J., Brown A., Bowden R.et al. .. Comparison of next-generation sequencing technologies for comprehensive assessment of full-length hepatitis C viral genomes. J. Clin. Microbiol. 2016; 54:2470–2484. - PMC - PubMed
MeSH terms
Substances
Grants and funding
- Finnish Medical Foundation
- Alfred Kordelin Foundation
- Finnish Cultural Foundation
- Orion Research Foundation
- Emil Aaltonen Foundation
- Finnish-Norwegian Medical Foundation
- Instrumentarium Science Foundation
- Biomedicum Helsinki Foundation
- Paulo Foundation
- Maud Kuistila Memorial Foundation
- Juhani Aho Foundation for Medical Research
- Helsingin ja Uudenmaan Sairaanhoitopiiri
- Finska Läkäresällskapet
- Magnus Ehrnrooth Foundation
- Jane and Aatos Erkko Foundation
- Sigrid Jusélius Foundation
- Medicinska Understödsföreningen Liv och Hälsa
- Finnish Society of Sciences and Letters
- Kone Foundation
- Academy of Finland
- Fundação para a Ciência e a Tecnologia
- University of Helsinki
LinkOut - more resources
Full Text Sources

