CXCL13 promotes broad immune responses induced by circular RNA vaccines
- PMID: 39436660
- PMCID: PMC11536096
- DOI: 10.1073/pnas.2406434121
CXCL13 promotes broad immune responses induced by circular RNA vaccines
Abstract
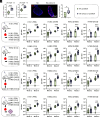
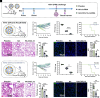
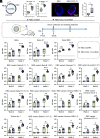
Antibody responses induced by current vaccines for influenza and SARS-CoV-2 often lack robust cross-reactivity. As hubs where diverse immune cells converge and interact, the alterations in the immune microenvironment within lymph nodes (LNs) are intricately linked to immune responses. Herein, we designed a lipid nanoparticle (LNP) loaded with circular RNA (circRNA) and targeted to LNs, in which CXCL13 was directly integrated into antigen-encoding circRNA strands. We demonstrated that CXCL13 alters the transcriptomic profiles of LNs, especially the upregulation of IL-21 and IL-4. Meanwhile, CXCL13 promotes the formation of germinal center and elicits robust antigen-specific T cell responses. With the codelivery of CXCL13 and the antigen, CXCL13 enhances cross-reactive antibodies against influenza virus and SARS-CoV-2, achieving protection against both homologous and heterologous influenza virus challenges in a mouse model. Notably, the targeted modification of LNP surfaces with antibodies helps address some of the challenges associated with lyophilized LNP vaccines, which is crucial for the long-term storage of LNP-circRNA vaccines. Overall, the circRNA-based antigen-CXCL13 coexpression system developed herein provides a simple and robust platform that enhances the magnitude and breadth of antibody responses against multiple viral glycoproteins, highlighting the potential utility of CXCL13 in inducing broad immune responses.
Keywords: CXCL13; SARS-CoV-2; broadly cross-reactive antibodies; circRNA vaccine; influenza virus.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures



References
-
- Aljabali A. A. A., et al. , Current state of, prospects for, and obstacles to mRNA vaccine development. Drug Discov. Today 28, 103458 (2023). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

