Distinct mechanisms drive divergent phenotypes in hypertrophic and dilated cardiomyopathy-associated TPM1 variants
- PMID: 39436707
- PMCID: PMC11645150
- DOI: 10.1172/JCI179135
Distinct mechanisms drive divergent phenotypes in hypertrophic and dilated cardiomyopathy-associated TPM1 variants
Abstract
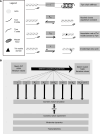
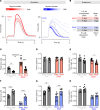
Heritable forms of hypertrophic cardiomyopathy (HCM) and dilated cardiomyopathy (DCM) represent starkly diverging clinical phenotypes, yet may be caused by mutations to the same sarcomeric protein. The precise mechanisms by which point mutations within the same gene bring about phenotypic diversity remain unclear. Our objective was to develop a mechanistic explanation of diverging phenotypes in two TPM1 mutations, E62Q (HCM) and E54K (DCM). Drawing on data from the literature and experiments with stem cell-derived cardiomyocytes expressing the TPM1 mutations of interest, we constructed computational simulations that provide plausible explanations of the distinct muscle contractility caused by each variant. In E62Q, increased calcium sensitivity and hypercontractility was explained most accurately by a reduction in effective molecular stiffness of tropomyosin and alterations in its interactions with the actin thin filament that favor the "closed" regulatory state. By contrast, the E54K mutation appeared to act via long-range allosteric interactions to increase the association rate of the C-terminal troponin I mobile domain to tropomyosin/actin. These mutation-linked molecular events produced diverging alterations in gene expression that can be observed in human engineered heart tissues. Modulators of myosin activity confirmed our proposed mechanisms by rescuing normal contractile behavior in accordance with predictions.
Keywords: Cardiology; Cardiovascular disease.
Conflict of interest statement
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

