Combined HDAC8 and checkpoint kinase inhibition induces tumor-selective synthetic lethality in preclinical models
- PMID: 39436709
- PMCID: PMC11601943
- DOI: 10.1172/JCI165448
Combined HDAC8 and checkpoint kinase inhibition induces tumor-selective synthetic lethality in preclinical models
Abstract
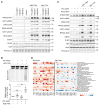
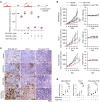
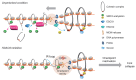
The elevated level of replication stress is an intrinsic characteristic of cancer cells. Targeting the mechanisms that maintain genome stability to further increase replication stress and thus induce severe genome instability has become a promising approach for cancer treatment. Here, we identify histone deacetylase 8 (HDAC8) as a drug target whose inactivation synergized with the inhibition of checkpoint kinases to elicit substantial replication stress and compromise genome integrity selectively in cancer cells. We showed that simultaneous inhibition of HDAC8 and checkpoint kinases led to extensive replication fork collapse, irreversible cell-cycle arrest, and synergistic vulnerability in various cancer cells. The efficacy of the combination treatment was further validated in patient tumor-derived organoid (PDO) and xenograft mouse (PDX) models, providing important insights into patient-specific drug responses. Our data revealed that HDAC8 activity was essential for reducing the acetylation level of structural maintenance of chromosomes protein 3 (SMC3) ahead of replication forks and preventing R loop formation. HDAC8 inactivation resulted in slowed fork progression and checkpoint kinase activation. Our findings indicate that HDAC8 guards the integrity of the replicating genome, and the cancer-specific synthetic lethality between HDAC8 and checkpoint kinases provides a promising replication stress-targeting strategy for treating a broad range of cancers.
Keywords: Cancer; Cell biology; Cell cycle; Genetic instability; Therapeutics.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

