MOGAT3-mediated DAG accumulation drives acquired resistance to anti-BRAF/anti-EGFR therapy in BRAFV600E-mutant metastatic colorectal cancer
- PMID: 39436710
- PMCID: PMC11645146
- DOI: 10.1172/JCI182217
MOGAT3-mediated DAG accumulation drives acquired resistance to anti-BRAF/anti-EGFR therapy in BRAFV600E-mutant metastatic colorectal cancer
Abstract
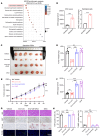
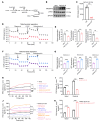
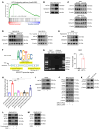
BRAFV600E-mutant metastatic colorectal cancer (mCRC) is associated with poor prognosis. The combination of anti-BRAF/anti-EGFR (encorafenib/cetuximab) treatment for patients with BRAFV600E-mutant mCRC improves clinical benefits; unfortunately, inevitable acquired resistance limits the treatment outcome, and the mechanism has not been validated. Here, we discovered that monoacylglycerol O-acyltransferase 3-mediated (MOGAT3-mediated) diacylglycerol (DAG) accumulation contributed to acquired resistance to encorafenib/cetuximab by dissecting a BRAFV600E-mutant mCRC patient-derived xenograft (PDX) model exposed to encorafenib/cetuximab administration. Mechanistically, the upregulated MOGAT3 promoted DAG synthesis and reduced fatty acid oxidation-promoting DAG accumulation and activated PKCα/CRAF/MEK/ERK signaling, driving acquired resistance. Resistance-induced hypoxia promoted MOGAT3 transcriptional elevation; simultaneously, MOGAT3-mediated DAG accumulation increased HIF1A expression at the translation level through PKCα/CRAF/eIF4E activation, strengthening the resistance status. Intriguingly, reducing intratumoral DAG with fenofibrate or PF-06471553 restored the antitumor efficacy of encorafenib/cetuximab in resistant BRAFV600E-mutant mCRC, which interrupted PKCα/CRAF/MEK/ERK signaling. These findings reveal the critical role of the metabolite DAG as a modulator of encorafenib/cetuximab efficacy in BRAFV600E-mutant mCRC, suggesting that fenofibrate might prove beneficial for resistant BRAFV600E-mutant mCRC patients.
Keywords: Colorectal cancer; Drug therapy; Gastroenterology; Therapeutics.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

