RRM2 inhibition alters cell cycle through ATM/Rb/E2F1 pathway in atypical teratoid rhabdoid tumor
- PMID: 39437704
- PMCID: PMC11536058
- DOI: 10.1016/j.neo.2024.101075
RRM2 inhibition alters cell cycle through ATM/Rb/E2F1 pathway in atypical teratoid rhabdoid tumor
Abstract
Background: Atypical teratoid rhabdoid tumor (ATRT) is an aggressive brain tumor that mainly affects young children. Our recent study reported a promising therapeutic strategy to trigger DNA damage, impede homologous recombination repair, and induce apoptosis in ATRT cells by targeting ribonucleotide reductase regulatory subunit M2 (RRM2). COH29, an inhibitor of RRM2, effectively reduced tumor growth and prolonged survival in vivo. Herein, we explored the underlying mechanisms controlling these functions to improve the clinical applicability of COH29 in ATRT.
Methods: Molecular profiling of ATRT patients and COH29-treated cells was analyzed to identify the specific signaling pathways, followed by validation using a knockdown system, flow cytometry, q-PCR, and western blot.
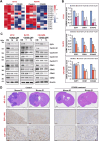
Results: Elevated E2F1 and its signaling pathway were correlated with poor prognosis. RRM2 inhibition induced DNA damage and activated ATM, which reduced Rb phosphorylation to promote Rb-E2F1 interaction and hindered E2F1 functions. E2F1 activity suppression led to decreased E2F1-dependent target expressions, causing cell cycle arrest in the G1 phase, decreased S phase cells, and blocked DNA damage repair.
Conclusion: Our study highlights the role of ATM/Rb/E2F1 pathway in controlling cell cycle arrest and apoptosis in response to RRM2 inhibition-induced DNA damage. This provides insight into the therapeutic benefits of COH29 and suggests targeting this pathway as a potential treatment for ATRT.
Keywords: ATM/Rb/E2F1; ATRT; COH29; Cell cycle; RRM2.
Copyright © 2024. Published by Elsevier Inc.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures






References
-
- Hilden JM, Meerbaum S, Burger P, Finlay J, Janss A, Scheithauer BW, et al. Central nervous system atypical teratoid/rhabdoid tumor: results of therapy in children enrolled in a registry. J. Clin. Oncol. 2004;22(14):2877–2884. - PubMed
-
- Matthews HK, Bertoli C, de Bruin RAM. Cell cycle control in cancer. Nat. Rev. Mol. Cell Biol. 2022;23(1):74–88. - PubMed
-
- Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 2008;9(4):297–308. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

