CENCAT enables immunometabolic profiling by measuring protein synthesis via bioorthogonal noncanonical amino acid tagging
- PMID: 39437716
- PMCID: PMC11573747
- DOI: 10.1016/j.crmeth.2024.100883
CENCAT enables immunometabolic profiling by measuring protein synthesis via bioorthogonal noncanonical amino acid tagging
Abstract
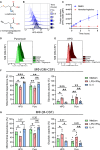
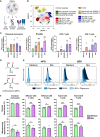
Cellular energy metabolism significantly contributes to immune cell function. To further advance immunometabolic research, novel methods to study the metabolism of immune cells in complex samples are required. Here, we introduce CENCAT (cellular energetics through noncanonical amino acid tagging). This technique utilizes click labeling of alkyne-bearing noncanonical amino acids to measure protein synthesis inhibition as a proxy for metabolic activity. CENCAT successfully reproduced known metabolic signatures of lipopolysaccharide (LPS)/interferon (IFN)γ and interleukin (IL)-4 activation in human primary macrophages. Application of CENCAT in peripheral blood mononuclear cells revealed diverse metabolic rewiring upon stimulation with different activators. Finally, CENCAT was used to analyze the cellular metabolism of murine tissue-resident immune cells from various organs. Tissue-specific clustering was observed based on metabolic profiles, likely driven by microenvironmental priming. In conclusion, CENCAT offers valuable insights into immune cell metabolic responses, presenting a powerful platform for studying cellular metabolism in complex samples and tissues in both humans and mice.
Keywords: CP: Immunology; CP: Metabolism; OXPHOS; SCENITH; energy metabolism; glycolysis; immunometabolism.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
-
- Shirai T., Nazarewicz R.R., Wallis B.B., Yanes R.E., Watanabe R., Hilhorst M., Tian L., Harrison D.G., Giacomini J.C., Assimes T.L., et al. The glycolytic enzyme PKM2 bridges metabolic and inflammatory dysfunction in coronary artery disease. J. Exp. Med. 2016;213:337–354. doi: 10.1084/jem.20150900. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

