Mechanism-free repurposing of drugs for C9orf72-related ALS/FTD using large-scale genomic data
- PMID: 39437787
- PMCID: PMC11605688
- DOI: 10.1016/j.xgen.2024.100679
Mechanism-free repurposing of drugs for C9orf72-related ALS/FTD using large-scale genomic data
Abstract
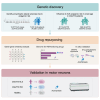
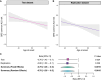
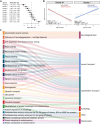
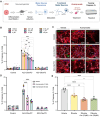
Repeat expansions in the C9orf72 gene are the most common genetic cause of (ALS) and frontotemporal dementia (FTD). Like other genetic forms of neurodegeneration, pinpointing the precise mechanism(s) by which this mutation leads to neuronal death remains elusive, and this lack of knowledge hampers the development of therapy for C9orf72-related disease. We used an agnostic approach based on genomic data (n = 41,273 ALS and healthy samples, and n = 1,516 C9orf72 carriers) to overcome these bottlenecks. Our drug-repurposing screen, based on gene- and expression-pattern matching and information about the genetic variants influencing onset age among C9orf72 carriers, identified acamprosate, a γ-aminobutyric acid analog, as a potentially repurposable treatment for patients carrying C9orf72 repeat expansions. We validated its neuroprotective effect in cell models and showed comparable efficacy to riluzole, the current standard of care. Our work highlights the potential value of genomics in repurposing drugs in situations where the underlying pathomechanisms are inherently complex. VIDEO ABSTRACT.
Keywords: C9orf72; acamprosate; age at onset; amyotrophic lateral sclerosis; drug repurposing; frontotemporal dementia; translation.
Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests B.J.T. holds patents on clinical testing and therapeutic intervention for the hexanucleotide repeat expansion of C9orf72.
Figures



References
-
- Renton A.E., Majounie E., Waite A., Simón-Sánchez J., Rollinson S., Gibbs J.R., Schymick J.C., Laaksovirta H., van Swieten J.C., Myllykangas L., et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. - DOI - PMC - PubMed
-
- DeJesus-Hernandez M., Mackenzie I.R., Boeve B.F., Boxer A.L., Baker M., Rutherford N.J., Nicholson A.M., Finch N.A., Flynn H., Adamson J., et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. - DOI - PMC - PubMed
-
- Majounie E., Renton A.E., Mok K., Dopper E.G.P., Waite A., Rollinson S., Chiò A., Restagno G., Nicolaou N., Simon-Sanchez J., et al. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 2012;11:323–330. doi: 10.1016/S1474-4422(12)70043-1. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

