IQGAP3 signalling mediates intratumoral functional heterogeneity to enhance malignant growth
- PMID: 39438124
- PMCID: PMC11874294
- DOI: 10.1136/gutjnl-2023-330390
IQGAP3 signalling mediates intratumoral functional heterogeneity to enhance malignant growth
Abstract
Background: The elevation of IQGAP3 expression in diverse cancers indicates a key role for IQGAP3 in carcinogenesis. Although IQGAP3 was established as a proliferating stomach stem cell factor and a regulator of the RAS-ERK pathway, how it drives cancer growth remains unclear.
Objective: We define the function of IQGAP3 in gastric cancer (GC) development and progression.
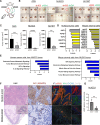
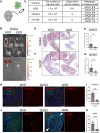
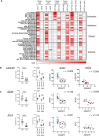
Design: We studied the phenotypic changes caused by IQGAP3 knockdown in three molecularly diverse GC cell lines by RNA-sequencing. In vivo tumorigenesis and lung metastasis assays corroborated IQGAP3 as a mediator of oncogenic signalling. Spatial analysis was performed to evaluate the intratumoral transcriptional and functional differences between control tumours and IQGAP3 knockdown tumours.
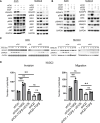
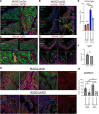
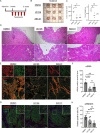
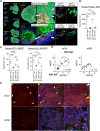
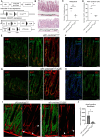
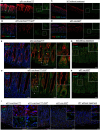
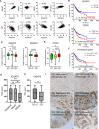
Results: Transcriptomic profiling showed that IQGAP3 inhibition attenuates signal transduction networks, such as KRAS signalling, via phosphorylation blockade. IQGAP3 knockdown was associated with significant inhibition of MEK/ERK signalling-associated growth factors, including TGFβ1, concomitant with gene signatures predictive of impaired tumour microenvironment formation and reduced metastatic potential. Xenografts involving IQGAP3 knockdown cells showed attenuated tumorigenesis and lung metastasis in immunodeficient mice. Accordingly, immunofluorescence staining revealed significant reductions of TGFβ/SMAD signalling and αSMA-positive stromal cells; digital spatial analysis indicated that IQGAP3 is indispensable for the formation of two phenotypically diverse cell subpopulations, which played crucial but distinct roles in promoting oncogenic functions.
Conclusion: IQGAP3 knockdown suppressed the RAS-TGFβ signalling crosstalk, leading to a significant reduction of the tumour microenvironment. In particular, IQGAP3 maintains functional heterogeneity of cancer cells to enhance malignant growth. IQGAP3 is thus a highly relevant therapy target in GC.
Keywords: carcinogenesis; signal transduction.
© Author(s) (or their employer(s)) 2025. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ Group.
Conflict of interest statement
Competing interests: None declared.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
