A splicing isoform of PD-1 promotes tumor progression as a potential immune checkpoint
- PMID: 39438489
- PMCID: PMC11496882
- DOI: 10.1038/s41467-024-53561-2
A splicing isoform of PD-1 promotes tumor progression as a potential immune checkpoint
Abstract
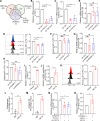
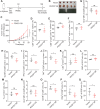
The immune checkpoint receptor, programmed cell death 1 (PD-1, encoded by PDCD1), mediates the immune escape of cancer, but whether PD-1 splicing isoforms contribute to this process is still unclear. Here, we identify an alternative splicing isoform of human PD-1, which carries a 28-base pairs extension retained from 5' region of intron 2 (PD-1^28), is expressed in peripheral T cells and tumor infiltrating lymphocytes. PD-1^28 expression is induced on T cells upon activation and is regulated by an RNA binding protein, TAF15. Functionally, PD-1^28 inhibits T cell proliferation, cytokine production, and tumor cell killing in vitro. In vivo, T cell-specific exogenous expression of PD-1^28 promotes tumor growth in both a syngeneic mouse tumor model and humanized NOG mice inoculated with human lung cancer cells. Our study thus demonstrates that PD-1^28 functions as an immune checkpoint, and may contribute to resistance to immune checkpoint blockade therapy.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

