A longitudinal single-cell atlas of anti-tumour necrosis factor treatment in inflammatory bowel disease
- PMID: 39438660
- PMCID: PMC11519010
- DOI: 10.1038/s41590-024-01994-8
A longitudinal single-cell atlas of anti-tumour necrosis factor treatment in inflammatory bowel disease
Abstract
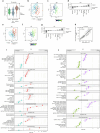
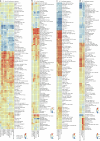
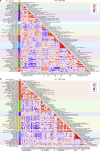
Precision medicine in immune-mediated inflammatory diseases (IMIDs) requires a cellular understanding of treatment response. We describe a therapeutic atlas for Crohn's disease (CD) and ulcerative colitis (UC) following adalimumab, an anti-tumour necrosis factor (anti-TNF) treatment. We generated ~1 million single-cell transcriptomes, organised into 109 cell states, from 216 gut biopsies (41 subjects), revealing disease-specific differences. A systems biology-spatial analysis identified granuloma signatures in CD and interferon (IFN)-response signatures localising to T cell aggregates and epithelial damage in CD and UC. Pretreatment differences in epithelial and myeloid compartments were associated with remission outcomes in both diseases. Longitudinal comparisons demonstrated disease progression in nonremission: myeloid and T cell perturbations in CD and increased multi-cellular IFN signalling in UC. IFN signalling was also observed in rheumatoid arthritis (RA) synovium with a lymphoid pathotype. Our therapeutic atlas represents the largest cellular census of perturbation with the most common biologic treatment, anti-TNF, across multiple inflammatory diseases.
© 2024. The Author(s).
Conflict of interest statement
T.T. has received research support from Celsius Therapeutics and consulting fees from Abbvie and ZuraBio. D.A. is an employee and shareholder of Novartis Pharma AG. This article reflects the authors’ personal opinions and not that of their employer. R.P. is employed by Scorpion Therapeutics and holds equity in Celsius Therapeutics. F.M.P. received research support from Roche and Janssen and consulting fees from GSK, Novartis and Genentech. H.H.U. received research support or consultancy fees from Janssen, Eli Lilly, UCB Pharma, BMS/Celgene, MiroBio, Mestag and OMass. A.F. has consulted for Janssen and Sonoma and has received research funding from BMS, Roche, UCB, Nascient, Mestag, GSK and Janssen. S.T. has received grants and research support from AbbVie, Buhlmann, Celgene, Celsius, ECCO, Helmsley Trust, IOIBD, Janssen, Lilly, Pfizer, Takeda, UKIERI, Vifor and Norman Collisson Foundation; consulting fees from AbbVie, ai4gi, Allergan, Amgen, Apexian, Arcturis, Arena, AstraZeneca, Bioclinica, Biogen, BMS, Buhlmann, Celgene, ChemoCentryx, Clario, Cosmo, Dynavax, Endpoint Health, Enterome, EQrX, Equillium, Ferring, Galapagos, Genentech/Roche, Gilead, GSK, Immunocore, Indigo, Janssen, Lilly, Mestag, Microbiotica, Novartis, Pfizer, Phesi, Protagonist, Sanofi, Satisfai, Sensyne Health, Sorriso, Syndermix, Takeda, Theravance, Topivert, UCB Pharma, VHsquared and Vifor; and speaker fees from AbbVie, Amgen, Biogen, BMS, Falk, Ferring, Janssen, Lilly, Pfizer and Takeda. C.D.B., M.B., M.C. and S.R. are founders of Mestag Therapeutics. M.F. received research support and consulting fees from Eli Lilly and Ono Pharmaceuticals. The remaining authors declare no competing interests.
Figures













References
-
- Brennan, F. M., Jackson, A., Chantry, D., Maini, R. & Feldmann, M. Inhibitory effect of TNF alpha antibodies on synovial cell interleukin-1 production in rheumatoid arthritis. Lancet2, 244–247 (1989). - PubMed
-
- Elliott, M. J. et al. Randomised double-blind comparison of chimeric monoclonal antibody to tumour necrosis factor alpha (cA2) versus placebo in rheumatoid arthritis. Lancet344, 1105–1110 (1994). - PubMed
-
- Derkx, B. et al. Tumour-necrosis-factor antibody treatment in Crohn’s disease. Lancet342, 173–174 (1993). - PubMed
-
- Ding, N. S., Hart, A. & De Cruz, P. Systematic review: predicting and optimising response to anti-TNF therapy in Crohn’s disease: algorithm for practical management. Aliment Pharm. Ther.43, 30–51 (2016). - PubMed
-
- Weinblatt, M. E. et al. Adalimumab, a fully human anti-tumor necrosis factor alpha monoclonal antibody, for the treatment of rheumatoid arthritis in patients taking concomitant methotrexate: the ARMADA trial. Arthritis Rheum.48, 35–45 (2003). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

