Integrating D4Z4 methylation analysis into clinical practice: improvement of FSHD molecular diagnosis through distinct thresholds for 4qA/4qA and 4qA/4qB patients
- PMID: 39438900
- PMCID: PMC11520157
- DOI: 10.1186/s13148-024-01747-2
Integrating D4Z4 methylation analysis into clinical practice: improvement of FSHD molecular diagnosis through distinct thresholds for 4qA/4qA and 4qA/4qB patients
Abstract
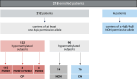
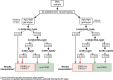
Background: Facioscapulohumeral dystrophy (FSHD) is a myopathy characterized by the loss of repressive epigenetic features affecting the D4Z4 locus (4q35). The assessment of DNA methylation at two regions (DUX4-PAS and DR1) of D4Z4 locus proved to be an effective method to detect epigenetic signatures compatible with FSHD. The present study aims at validating the employment of this method into clinical practice and improving the protocol by refining the classification thresholds of 4qA/4qA patients. To this purpose, 218 subjects with clinical suspicion of FSHD collected in 2022-2023 were analyzed. Each participant underwent in parallel the traditional FSHD molecular testing (D4Z4 sizing) and the proposed methylation assay. The results provided by both analyses were compared to evaluate the concordance and calculate the performance metrics of the methylation test.
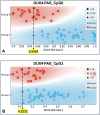
Results: Among the 218 subjects, the 4q variant type distribution was 54% 4qA/4qA, 43% 4qA/4qB and 3% 4qB/4qB. The methylation analysis was performed only on carriers of at least one 4qA allele. After refining the classification threshold, the test reached the following performance metrics: sensitivity = 0.90, specificity = 1.00 and accuracy = 0.93. These results confirmed the effectiveness of the methylation assay in identifying patients with genetic signature compatible with FSHD1 and FSHD2 based on their DUX4-PAS and DR1 profile, respectively. The methylation data were also evaluated with respect to the clinical information.
Conclusions: The study confirmed the ability of the method to accurately identify methylation profiles compatible with FSHD genetic signatures considering the 4q genotype. Moreover, the test allows the detection of hypomethylated profiles in asymptomatic patients, suggesting its potential application in identifying preclinical conditions in patients with positive family history and FSHD genetic signatures. Furthermore, the present work emphasizes the importance of interpreting methylation profiles considering the patients' clinical data.
Keywords: D4Z4; DNA methylation; Epigenetic biomarker; FSHD; FSHD diagnosis; FSHD signature; Neuromuscular diseases.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

