Golgiphagy: a novel selective autophagy to the fore
- PMID: 39438975
- PMCID: PMC11495120
- DOI: 10.1186/s13578-024-01311-8
Golgiphagy: a novel selective autophagy to the fore
Abstract
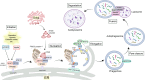
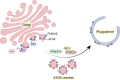
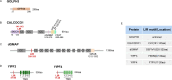
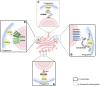
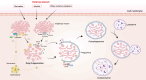
The Golgi apparatus is the central hub of the cellular endocrine pathway and plays a crucial role in processing, transporting, and sorting proteins and lipids. Simultaneously, it is a highly dynamic organelle susceptible to degradation or fragmentation under various physiological or pathological conditions, potentially contributing to the development of numerous human diseases. Autophagy serves as a vital pathway for eukaryotes to manage intracellular and extracellular stress and maintain homeostasis by targeting damaged or redundant organelles for removal. Recent research has revealed that autophagy mechanisms can specifically degrade Golgi components, known as Golgiphagy. This review summarizes recent findings on Golgiphagy while also addressing unanswered questions regarding its mechanisms and regulation, aiming to advance our understanding of the role of Golgiphagy in human disease.
Keywords: Autophagy; Golgi apparatus; Golgi fragmentation; Golgiphagy; Receptor.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Bentivoglio M. 1898: the golgi apparatus emerges from nerve cells. Trends Neurosci. 1998;21:195–200. - PubMed
-
- Short B, Barr FA. The golgi apparatus. Curr Biol CB. 2000;10:R583–585. - PubMed
-
- Potelle S, Klein A, Foulquier F. Golgi post-translational modifications and associated diseases. J Inherit Metab Dis. 2015;38:741–51. - PubMed
-
- De Matteis MA, Luini A. Exiting the golgi complex. Nat Rev Mol Cell Biol. 2008;9:273–84. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

