Assessing the impact of deep-learning assistance on the histopathological diagnosis of serous tubal intraepithelial carcinoma (STIC) in fallopian tubes
- PMID: 39439213
- PMCID: PMC11496567
- DOI: 10.1002/2056-4538.70006
Assessing the impact of deep-learning assistance on the histopathological diagnosis of serous tubal intraepithelial carcinoma (STIC) in fallopian tubes
Abstract
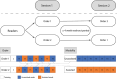
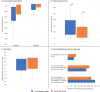
In recent years, it has become clear that artificial intelligence (AI) models can achieve high accuracy in specific pathology-related tasks. An example is our deep-learning model, designed to automatically detect serous tubal intraepithelial carcinoma (STIC), the precursor lesion to high-grade serous ovarian carcinoma, found in the fallopian tube. However, the standalone performance of a model is insufficient to determine its value in the diagnostic setting. To evaluate the impact of the use of this model on pathologists' performance, we set up a fully crossed multireader, multicase study, in which 26 participants, from 11 countries, reviewed 100 digitalized H&E-stained slides of fallopian tubes (30 cases/70 controls) with and without AI assistance, with a washout period between the sessions. We evaluated the effect of the deep-learning model on accuracy, slide review time and (subjectively perceived) diagnostic certainty, using mixed-models analysis. With AI assistance, we found a significant increase in accuracy (p < 0.01) whereby the average sensitivity increased from 82% to 93%. Further, there was a significant 44 s (32%) reduction in slide review time (p < 0.01). The level of certainty that the participants felt versus their own assessment also significantly increased, by 0.24 on a 10-point scale (p < 0.01). In conclusion, we found that, in a diverse group of pathologists and pathology residents, AI support resulted in a significant improvement in the accuracy of STIC diagnosis and was coupled with a substantial reduction in slide review time. This model has the potential to provide meaningful support to pathologists in the diagnosis of STIC, ultimately streamlining and optimizing the overall diagnostic process.
Keywords: STIC; artificial intelligence; computational pathology; deep learning; high‐grade serous carcinoma; histopathology; serous tubal intraepithelial carcinoma.
© 2024 The Author(s). The Journal of Pathology: Clinical Research published by The Pathological Society of Great Britain and Ireland and John Wiley & Sons Ltd.
Figures



References
-
- Pallua JD, Brunner A, Zelger B, et al. The future of pathology is digital. Pathol Res Pract 2020; 216: 153040. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

