Genetic signatures of AKT1 variants associated with worse COVID-19 outcomes - a multicentric observational study
- PMID: 39439795
- PMCID: PMC11493623
- DOI: 10.3389/fimmu.2024.1422349
Genetic signatures of AKT1 variants associated with worse COVID-19 outcomes - a multicentric observational study
Abstract
Introduction: The COVID-19, triggered by the SARS-CoV-2 virus, has varied clinical manifestations, ranging from mild cases to severe forms such as fatal pneumonia and acute respiratory distress syndrome (ARDS). Disease severity is influenced by an exacerbated immune response, characterized by high pro-inflammatory cytokine levels. Inhibition of AKT can potentially suppress pathological inflammation, cytokine storm and platelet activation associated with COVID-19. In this study, we aimed to investigate the rs2494746 and rs1130214 variants in the AKT1 gene associated with severe COVID-19 outcomes.
Methods: Peripheral blood samples and sociodemographic data from 508 individuals with COVID-19, measuring plasma cytokine concentrations using ELISA and genotyped the AKT1 variants.
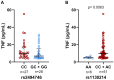
Results: The rs2494746-C allele was associated with severity, ICU admission, and death from COVID-19. The C allele at rs1130214 was linked to increased TNF and D-dimer levels. Moreover, both variants exhibited an increased cumulative risk of disease severity, ICU admission, and mortality caused by COVID-19. In the predictive analysis, the rs2494746 obtained an accuracy of 71%, suggesting a high probability of the test determining the severity of the disease.
Discussion: Our findings contribute to understanding the influence of the AKT1 gene variants on the immunological damage in individuals infected with SARS-CoV-2.
Keywords: AKT1; COVID-19; immunogenetics; polymorphism; severity.
Copyright © 2024 de Almeida, Tosta, Pena, Silva, Reis-Goes, Silva, Cruz, Silva, de Araújo, Rodrigues, Oliveira, Figueiredo, Vaz, Montaño-Castellón, Santana, Torres, Beltrão, Carneiro, Campos, Brites, Fortuna, Figueiredo, Trindade, Ramos and Costa.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

