A novel m7G-related miRNA prognostic signature for predicting clinical outcome and immune microenvironment in colon cancer
- PMID: 39440054
- PMCID: PMC11493006
- DOI: 10.7150/jca.99173
A novel m7G-related miRNA prognostic signature for predicting clinical outcome and immune microenvironment in colon cancer
Abstract
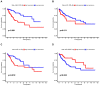
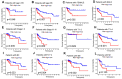
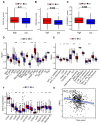
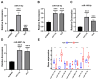
Background: Colon cancer (CC) is a highly prevalent malignancy worldwide, characterized by elevated mortality rates and poor prognosis. N7-methylguanosine (m7G) methylation is an emerging RNA modification type and involved in the development of many tumors. Despite this, the correlation between m7G-related miRNAs and CC remains to be elucidated. This research aimed to investigate the clinical significance of m7G-related miRNAs in predicting both the prognosis and tumor microenvironment (TME) of CC. Method: We retrieved transcriptome data and associated clinical information from a publicly accessible database. Using univariate Cox and LASSO regression analyses, we established a signature of m7G-related miRNAs. Additionally, we used CIBERSORT and ssGSEA algorithms to explore the association between the prognostic risk score and the TME in CC patients. By considering the risk signature and immune infiltration, we identified differentially expressed genes that contribute to the prognosis of CC. Finally, the expression patterns of prognostic miRNAs were verified using quantitative reverse transcriptase PCR (qRT-PCR) in cell lines. Results: We constructed a prognostic risk signature based on seven m7G-related miRNAs (miR-136-5p, miR-6887-3p, miR-195-5p, miR-149-3p, miR-4433a-5p, miR-31-5p, and miR-129-2-3p). Subsequently, we observed remarkable differences in patient outcomes between the high- and low-risk groups. The area under the curve (AUC) for 1-, 3-, and 5-year survivals in the ROC curve were 0.735, 0.707, and 0.632, respectively. Furthermore, our results showed that the risk score can serve as an independent prognostic biomarker for overall survival prediction. In terms of immune analysis, the results revealed a significant association between the risk signature and immune infiltration, as well as immune checkpoint expression. Finally, our study showed that CCDC160 and RLN3 is the gene most relevant to immune cells and function in CC. Conclusion: Our study conducted a comprehensive and systematic analysis of m7G-associated miRNAs to construct prognostic profiles of CC. We developed a prognostic risk model based on m7G-miRNAs, with the resulting risk scores demonstrating considerable potential as prognostic biomarkers. These findings provide substantial evidence for the critical role of m7G-related miRNAs in colon cancer and may offer new immunotherapeutic targets for patients with this disease.
Keywords: N7-methylguanosine; colon cancer; immune microenvironment; microRNA; prognostic signature.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures








References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209–249. - PubMed
-
- Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73(1):17–48. - PubMed
-
- O'Sullivan DE, Sutherland RL, Town S, Chow K, Fan J, Forbes N, Heitman SJ, Hilsden RJ, Brenner DR. Risk Factors for Early-Onset Colorectal Cancer: A Systematic Review and Meta-analysis. Clin Gastroenterol Hepatol. 2022;20(6):1229–1240. e5. - PubMed
-
- Biller LH, Schrag D. Diagnosis and Treatment of Metastatic Colorectal Cancer: A Review. Jama. 2021;325(7):669–685. - PubMed
LinkOut - more resources
Full Text Sources

