A Strategic Framework of SARS-CoV-2 Genomic Surveillance in Bangladesh
- PMID: 39440811
- PMCID: PMC11497173
- DOI: 10.1111/irv.70019
A Strategic Framework of SARS-CoV-2 Genomic Surveillance in Bangladesh
Abstract
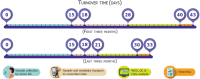
Background: The global pandemic caused by SARS-CoV-2 has underlined the significance of strict genomic surveillance to track virus evolution and the possible emergence of new variants, particularly in densely populated countries like Bangladesh. This study outlines a strategic framework of genomic surveillance to track the evolution of the virus in Bangladesh between June 2021 and December 2022 through the National SARS-CoV-2 Variant Surveillance (NSVS) program involving collaboration across 4 major institutes and 13 hospitals nationwide.
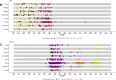
Methods: We aimed to capture the variants of SARS-CoV-2 throughout the country utilizing standardized procedures, modern sequencing technology, and stringent quality control, promoting data accuracy and the timely detection of new variants of concern. We sequenced over 2200 genomes, documenting the prevalence of the Delta variant initially, followed by the emergence of Omicron variants BA.1, BA.2, BA.5, and XBB, each affecting transmission rates and vaccine efficacy differently.
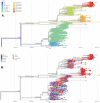
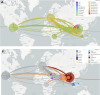
Results: The clinical manifestations of the variants differed, with some symptoms occurring more frequently in Delta cases and vice versa. Vaccinated individuals were more affected by Omicron cases compared to Delta cases. These variants were responsible for two major COVID-19 waves in the country, each with significant clinical effects. Phylogenetic analyses placed the local SARS-CoV-2 variants within a global context, indicating the Delta variant likely entered from India and Omicron from Europe.
Conclusion: This research highlights the significance of collaborative surveillance strategies for guiding public health choices and the critical role of genomic analysis in monitoring virus evolution, shaping targeted pandemic responses. Bangladesh's contributions significantly enhance global insight into COVID-19's genomic evolution.
Keywords: Bangladesh; COVID‐19; Delta; Omicron; SARS‐CoV‐2; surveillance.
© 2024 The Author(s). Influenza and Other Respiratory Viruses Published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Comprehensive Analysis of SARS-CoV-2 Dynamics in Bangladesh: Infection Trends and Variants (2020-2023).Viruses. 2024 Aug 7;16(8):1263. doi: 10.3390/v16081263. Viruses. 2024. PMID: 39205237 Free PMC article.
-
Genomics-based approach for detection and characterization of SARS-CoV-2 co-infections and diverse viral populations.Microbiol Spectr. 2025 Jun 3;13(6):e0209224. doi: 10.1128/spectrum.02092-24. Epub 2025 May 1. Microbiol Spectr. 2025. PMID: 40310264 Free PMC article.
-
Baseline Sequencing Surveillance of Public Clinical Testing, Hospitals, and Community Wastewater Reveals Rapid Emergence of SARS-CoV-2 Omicron Variant of Concern in Arizona, USA.mBio. 2023 Feb 28;14(1):e0310122. doi: 10.1128/mbio.03101-22. Epub 2023 Jan 9. mBio. 2023. PMID: 36622143 Free PMC article.
-
SARS-CoV-2 Variants and COVID-19 in Bangladesh-Lessons Learned.Viruses. 2024 Jul 4;16(7):1077. doi: 10.3390/v16071077. Viruses. 2024. PMID: 39066238 Free PMC article. Review.
-
Dynamics of SARS-CoV-2 variants in West Africa: Insights into genomic surveillance in resource-constrained settings.Infect Genet Evol. 2024 Nov;125:105681. doi: 10.1016/j.meegid.2024.105681. Epub 2024 Oct 20. Infect Genet Evol. 2024. PMID: 39437881 Free PMC article. Review.
References
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

