Evaluating the performance of multi-omics integration: a thyroid toxicity case study
- PMID: 39441382
- PMCID: PMC11742338
- DOI: 10.1007/s00204-024-03876-2
Evaluating the performance of multi-omics integration: a thyroid toxicity case study
Abstract
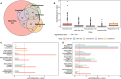
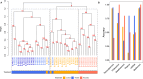
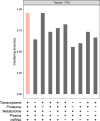
Multi-omics data integration has been repeatedly discussed as the way forward to more comprehensively cover the molecular responses of cells or organisms to chemical exposure in systems toxicology and regulatory risk assessment. In Canzler et al. (Arch Toxicol 94(2):371-388. https://doi.org/10.1007/s00204-020-02656-y ), we reviewed the state of the art in applying multi-omics approaches in toxicological research and chemical risk assessment. We developed best practices for the experimental design of multi-omics studies, omics data acquisition, and subsequent omics data integration. We found that multi-omics data sets for toxicological research questions were generally rare, with no data sets comprising more than two omics layers adhering to these best practices. Due to these limitations, we could not fully assess the benefits of different data integration approaches or quantitatively evaluate the contribution of various omics layers for toxicological research questions. Here, we report on a multi-omics study on thyroid toxicity that we conducted in compliance with these best practices. We induced direct and indirect thyroid toxicity through Propylthiouracil (PTU) and Phenytoin, respectively, in a 28-day plus 14-day recovery oral rat toxicity study. We collected clinical and histopathological data and six omics layers, including the long and short transcriptome, proteome, phosphoproteome, and metabolome from plasma, thyroid, and liver. We demonstrate that the multi-omics approach is superior to single-omics in detecting responses at the regulatory pathway level. We also show how combining omics data with clinical and histopathological parameters facilitates the interpretation of the data. Furthermore, we illustrate how multi-omics integration can hint at the involvement of non-coding RNAs in post-transcriptional regulation. Also, we show that multi-omics facilitates grouping, and we assess how much information individual and combinations of omics layers contribute to this approach.
Keywords: Chemical exposure; Data integration; Multi-omics; Risk assessment; Toxicology.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Conflict of interest: The authors declare that they have no potential conflict of interest.
Figures


References
-
- Amorim MJB, Peijnenburg W, Greco D, Saarimäki LA, Dumit VI, Bahl A, Haase A, Tran L, Hackermüller J, Canzler S, Scott-Fordsmand JJ (2023) Systems toxicology to advance human and environmental hazard assessment: a roadmap for advanced materials. Nano Today 48:101735. 10.1016/j.nantod.2022.101735 (https://www.sciencedirect.com/science/article/pii/S1748013222003632) - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

