A myeloid differentiation-like protein in partnership with Toll5 from the pest insect Spodoptera litura senses baculovirus infection
- PMID: 39441638
- PMCID: PMC11536157
- DOI: 10.1073/pnas.2415398121
A myeloid differentiation-like protein in partnership with Toll5 from the pest insect Spodoptera litura senses baculovirus infection
Abstract
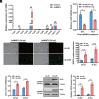
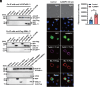
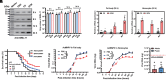
Many types of viruses infect insects and other arthropods. In contrast, little is known about how arthropods sense viruses, although several innate immune pathways including Toll have antiviral functions. Large DNA viruses in the family Baculoviridae are used to control a number of pest insects. Here, we studied Spodoptera litura and Autographa californica multiple nucleopolyhedrovirus (AcMNPV) to test the hypothesis that one or more myeloid differentiation-like (ML) proteins and Toll family members sense baculoviruses. We identified 11 ML and 12 Toll genes in the S. litura genome. A series of experiments indicated that S. litura ML protein 11 (SlML-11) binds the budded form of AcMNPV and partners with S. litura Toll5 (SlToll5). SlML-11 also bound sphingomyelin (SPM), which is a component of the virion envelope. Disabling SlML-11 and SlToll5 increased susceptibility to infection, while priming larvae with SPM reduced susceptibility as measured by increased survival to the adult stage and clearance of AcMNPV from individuals that emerged as adults. We conclude that SPM is a pathogen-associated molecular pattern molecule while SlML-11 and SlToll5 interact to function as a pattern recognition receptor that senses AcMNPV.
Keywords: immunity; insect; pattern recognition receptor; virus.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures



References
-
- Brennan J. J., Gilmore T. D., Evolutionary origins of toll-like receptor signaling. Mol. Biol. Evol. 35, 1576–1587 (2018). - PubMed
-
- Kawai T., Akira S., The role of pattern-recognition receptors in innate immunity: Update on toll-like receptors. Nat. Immunol. 11, 373–384 (2010). - PubMed
-
- Lemaitre B., Nicolas E., Michaut L., Reichhart J. M., Hoffmann J. A., The dorsoventral regulatory gene cassette spätzle/toll/cactus controls the potent antifungal response in Drosophila adults. Cell 86, 973–983 (1996). - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

