Repeated horizontal acquisition of lagriamide-producing symbionts in Lagriinae beetles
- PMID: 39441990
- PMCID: PMC11542224
- DOI: 10.1093/ismejo/wrae211
Repeated horizontal acquisition of lagriamide-producing symbionts in Lagriinae beetles
Abstract
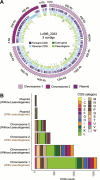
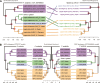
Microbial symbionts associate with multicellular organisms on a continuum from facultative associations to mutual codependency. In the oldest intracellular symbioses there is exclusive vertical symbiont transmission, and co-diversification of symbiotic partners over millions of years. Such symbionts often undergo genome reduction due to low effective population sizes, frequent population bottlenecks, and reduced purifying selection. Here, we describe multiple independent acquisition events of closely related defensive symbionts followed by genome erosion in a group of Lagriinae beetles. Previous work in Lagria villosa revealed the dominant genome-eroded symbiont of the genus Burkholderia produces the antifungal compound lagriamide, protecting the beetle's eggs and larvae from antagonistic fungi. Here, we use metagenomics to assemble 11 additional genomes of lagriamide-producing symbionts from 7 different host species within Lagriinae from 5 countries, to unravel the evolutionary history of this symbiotic relationship. In each host, we detected one dominant genome-eroded Burkholderia symbiont encoding the lagriamide biosynthetic gene cluster. However, we did not find evidence for host-symbiont co-diversification or for monophyly of the lagriamide-producing symbionts. Instead, our analyses support a single ancestral acquisition of the gene cluster followed by at least four independent symbiont acquisitions and subsequent genome erosion in each lineage. By contrast, a clade of plant-associated relatives retained large genomes but secondarily lost the lagriamide gene cluster. Our results, therefore, reveal a dynamic evolutionary history with multiple independent symbiont acquisitions characterized by a high degree of specificity and highlight the importance of the specialized metabolite lagriamide for the establishment and maintenance of this defensive symbiosis.
Keywords: Burkholderia; Lagriinae; biosynthetic gene cluster; chemical defense; lagriamide; metagenomics; secondary metabolism; symbiont replacement; symbiosis.
© The Author(s) 2024. Published by Oxford University Press on behalf of the International Society for Microbial Ecology.
Conflict of interest statement
The Kwan lab offers their metagenomic binning pipeline Autometa on the paid bioinformatics and computational platform BatchX in addition to distributing it through open source channels.
Figures




Update of
-
Repeated horizontal acquisition of lagriamide-producing symbionts in Lagriinae beetles.bioRxiv [Preprint]. 2024 Jul 9:2024.01.23.576914. doi: 10.1101/2024.01.23.576914. bioRxiv. 2024. Update in: ISME J. 2024 Jan 8;18(1):wrae211. doi: 10.1093/ismejo/wrae211. PMID: 39026795 Free PMC article. Updated. Preprint.
References
-
- Von Dohlen CD, Moran NA. Molecular data support a rapid radiation of aphids in the cretaceous and multiple origins of host alternation. Biol J Linn Soc Lond 2000;71:689–717. 10.1111/j.1095-8312.2000.tb01286.x - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

