Dehydrin Client Proteins Identified Using Phage Display Affinity Selected Libraries Processed With Paired-End Phage Sequencing
- PMID: 39442694
- PMCID: PMC11612773
- DOI: 10.1016/j.mcpro.2024.100867
Dehydrin Client Proteins Identified Using Phage Display Affinity Selected Libraries Processed With Paired-End Phage Sequencing
Abstract
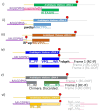
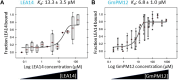
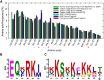
The late embryogenesis abundant proteins (LEAPs) are a class of noncatalytic, intrinsically disordered proteins with a malleable structure. Some LEAPs exhibit a protein and/or membrane binding capacity and LEAP binding to various targets has been positively correlated with abiotic stress tolerance. Regarding the LEAPs' presumptive role in protein protection, identifying client proteins (CtPs) to which LEAPs bind is one practicable means of revealing the mechanism by which they exert their function. To this end, we used phage display affinity selection to screen libraries derived from Arabidopsis thaliana seed mRNA with recombinant orthologous LEAPs from Arabidopsis and soybean (Glycine max). Subsequent high-throughput sequencing of DNA from affinity-purified phage was performed to characterize the entire subpopulation of phage retained by each LEAP ortholog. This entailed cataloging in-frame fusions, elimination of false positives, and aligning the hits on the CtP scaffold to reveal domains of respective CtPs that bound to orthologous LEAPs. This approach (paired-end phage sequencing) revealed a subpopulation of the proteome constituting the CtP repertoire in common between the two dehydrin orthologs (LEA14 and GmPm12) compared to bovine serum albumin (unrelated binding control). The veracity of LEAP:CtP binding for one of the CtPs (LEA14 and GmPM12 self-association) was independently assessed using temperature-related intensity change analysis. Moreover, LEAP:CtP interactions for four other CtPs were confirmed in planta using bimolecular fluorescence complementation assays. The results provide insights into the involvement of the dehydrin Y-segments and K-domains in protein binding.
Keywords: client proteins; late embryogenesis abundant proteins; paired-end sequencing; phage display; temperature related intensity change assay.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare no competing interests.
Figures









Similar articles
-
Identification of Late Embryogenesis Abundant (LEA) protein putative interactors using phage display.Int J Mol Sci. 2012;13(6):6582-6603. doi: 10.3390/ijms13066582. Epub 2012 May 29. Int J Mol Sci. 2012. PMID: 22837651 Free PMC article.
-
Uses of phage display in agriculture: sequence analysis and comparative modeling of late embryogenesis abundant client proteins suggest protein-nucleic acid binding functionality.Comput Math Methods Med. 2013;2013:470390. doi: 10.1155/2013/470390. Epub 2013 Jul 9. Comput Math Methods Med. 2013. PMID: 23956788 Free PMC article.
-
Wide screening of phage-displayed libraries identifies immune targets in planta.PLoS One. 2013;8(1):e54654. doi: 10.1371/journal.pone.0054654. Epub 2013 Jan 25. PLoS One. 2013. PMID: 23372747 Free PMC article.
-
Late Embryogenesis Abundant Protein-Client Protein Interactions.Plants (Basel). 2020 Jun 29;9(7):814. doi: 10.3390/plants9070814. Plants (Basel). 2020. PMID: 32610443 Free PMC article. Review.
-
Phage display biopanning and isolation of target-unrelated peptides: in search of nonspecific binders hidden in a combinatorial library.Amino Acids. 2016 Dec;48(12):2699-2716. doi: 10.1007/s00726-016-2329-6. Epub 2016 Sep 20. Amino Acids. 2016. PMID: 27650972 Review.
References
-
- Sallon S., Solowey E., Cohen Y., Korchinsky R., Egli M., Woodhatch I., et al. Germination, genetics, and growth of an ancient date seed. Science. 2008;320:1464. - PubMed
-
- Shen-Miller J., Mudgett M.B., Schopf J.W., Clarke S., Berger R. Exceptional seed longevity and robust growth: ancient sacred Lotus from China. Am. J. Bot. 1995;82:1367–1380.
-
- Tepfer D., Zalar A., Leach S. Survival of plant seeds, their UV screens, and nptII DNA for 18 months outside the International Space Station. Astrobiology. 2012;12:517–528. - PubMed
-
- Adhikari B.N., Wall D.H., Adams B.J. Effect of slow desiccation and freezing on gene transcription and stress survival of an Antarctic nematode. J. Exp. Biol. 2010;213:1803–1812. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

