Intra species dissection of phytophthora capsici resistance in black pepper
- PMID: 39442874
- PMCID: PMC12302342
- DOI: 10.1016/j.jare.2024.10.015
Intra species dissection of phytophthora capsici resistance in black pepper
Abstract
Introduction: Black pepper, a financially significant tropical crop, assumes a pivotal role in global agriculture for the major source of specie flavor. Nonetheless, the growth and productivity of black pepper face severe impediments due to the destructive pathogen Phytophthora capsici, ultimately resulting in black pepper blight. The dissecting for the genetic source of pathogen resistance for black pepper is beneficial for its global production. The genetic sources include the variations on gene coding sequences, transcription capabilities and epigenetic modifications, which exerts hierarchy of influences on plant defense against pathogen. However, the understanding of genetic source of disease resistance in black pepper remains limited.
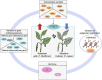
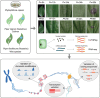
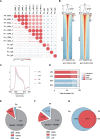
Methods: The wild species Piper flaviflorum (P. flaviflorum, Pf) is known for blight resistance, while the cultivated species P. nigrum is susceptible. To dissecting the genetic sources of pathogen resistance for black pepper, the chromatin modification on H3K4me3 and transcriptome of black pepper species were profiled for genome wide comparative studies, applied with CUT&Tag and RNA sequencing technologies.
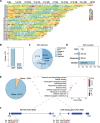
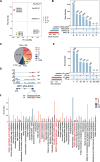
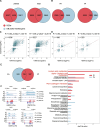
Results: The intraspecies difference between P. flaviflorum and P. nigrum on gene body region led to coding variations on 5137 genes, including 359 gene with biotic stress responses and regulation. P. flaviflorum exhibited a more comprehensive resistance response to Phytophthora capsici in terms of transcriptome features. The pathogen responsive transcribing was significant associated with histone modification mark of H3K4me3 in black pepper. The collective data on variations of sequence, transcription activity and chromatin structure lead to an exclusive jasmonic acid-responsive pathway for disease resistance in P. flaviflorum was revealed. This research provides a comprehensive frame work to identify the fine genetic source for pathogen resistance from wild species of black pepper.
Keywords: Black pepper; CUT&Tag; Disease resistance; Epigenetic; Jasmonic acid; Phytophthora capsici.
Copyright © 2024. Published by Elsevier B.V.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

References
-
- Butt M.S., Pasha I., Sultan M.T., Randhawa M.A., Saeed F., Ahmed A. Black pepper and health claims: A comprehensive treatise. Crit Rev Food Sci Nutr. 2013;53(9):875–886. - PubMed
-
- Nysanth N.S., Divya S., Nair CB., Anju A.B., Praveena R., Anith K.N. Biological control of foot rot (phytophthora capsici leonian) disease in black pepper (piper nigrum l.) with rhizospheric microorganisms. Rhizosphere. 2022;23:100578.
-
- Hausbeck M.K., Lamour K.H. Phytophthora capsici on vegetable crops: Research progress and management challenges. Plant Dis. 2004;88(12):1292–1303. - PubMed
-
- Zhao Y., Yang B., Xu H., Wu J., Xu Z., Wang Y. The phytophthora effector avh94 manipulates host jasmonic acid signaling to promote infection. J Integr Plant Biol. 2022;64(11):2199–2210. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

