RNA fold prediction by Monte Carlo in graph space and the statistical mechanics of tertiary interactions
- PMID: 39442981
- PMCID: PMC11648927
- DOI: 10.1261/rna.080216.124
RNA fold prediction by Monte Carlo in graph space and the statistical mechanics of tertiary interactions
Abstract
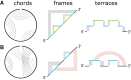
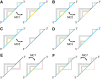
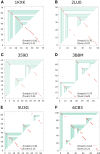
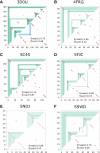
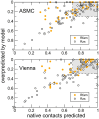
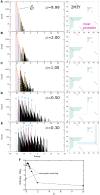
Using a graph representation of RNA structures, we have studied the ensembles of secondary and tertiary graphs of two sets of RNA with Monte Carlo simulations. The first consisted of 91 target ribozyme and riboswitch sequences of moderate lengths (<150 nt) having a variety of secondary, H-type pseudoknots and kissing loop interactions. The second set consisted of 71 more diverse sequences across many RNA families. Using a simple empirical energy model for tertiary interactions and only sequence information for each target as input, the simulations examined how tertiary interactions impact the statistical mechanics of the fold ensembles. The results show that the graphs proliferate enormously when tertiary interactions are possible, producing an entropic driving force for the ensemble to access folds having tertiary structures even though they are overall energetically unfavorable in the energy model. For each of the targets in the two test sets, we assessed the quality of the model and the simulations by examining how well the simulated structures were able to predict the native fold, and compared the results to fold predictions from ViennaRNA. Our model generated good or excellent predictions in a large majority of the targets. Overall, this method was able to produce predictions of comparable quality to Vienna, but it outperformed Vienna for structures with H-type pseudoknots. The results suggest that while tertiary interactions are predicated on real-space contacts, their impacts on the folded structure of RNA can be captured by graph space information for sequences of moderate lengths, using a simple tertiary energy model for the loops, the base pairs, and base stacks.
Keywords: Monte Carlo; RNA folding; fold prediction; graphs; secondary and tertiary structure.
© 2025 Phan and Mak.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
