Mitogen-activated protein kinase kinase kinase 1 facilitates the temozolomide resistance and migration of GBM via the MEK/ERK signalling
- PMID: 39443331
- PMCID: PMC11499072
- DOI: 10.1111/jcmm.70173
Mitogen-activated protein kinase kinase kinase 1 facilitates the temozolomide resistance and migration of GBM via the MEK/ERK signalling
Abstract
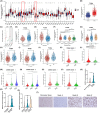
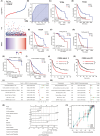
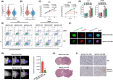
Mitogen-Activated Protein Kinase Kinase Kinase 1 (MAP3K1) is overexpressed in gliomas; however, its clinical significance, biological functions, and underlying molecular mechanisms remain unclear. Abnormal overexpression of MAP3K1 in glioma is strongly associated with unfavourable clinicopathological characteristics and disease progression. MAP3K1 could potentially serve as a reliable diagnostic and prognostic biomarker for glioma. MAP3K1 silencing suppressed the migration but had no effect on the proliferation and cell death of Glioblastoma Multiforme (GBM) cells. MAP3K1 knockdown exacerbated the temozolomide (TMZ) induced inhibition of glioma cell proliferation and death of GBM cells. In addition, MAP3K1 knockdown combined with TMZ treatment significantly inhibited the growth and increased cell death in organoids derived from GBM patients. MAP3K1 knockdown reversed TMZ resistance of GBM in intracranial glioma model. In terms of molecular mechanisms, the phosphorylation level of ERK was significantly decreased by MAP3K1 silencing. No significant change in the JNK pathway was found in MAP3K1-silenced GBM cells. Inhibition of ERK phosphorylation suppressed the migration and enhanced the TMZ sensibility of GBM cells. MAP3K1 was correlated with the immune infiltration in glioma. MAP3K1 could facilitate the migration and TMZ resistance of GBM cells through MEK/ERK signalling.
Keywords: MAP3K1; MEK/ERK; TMZ resistance; glioma; migration; patients‐derived organoids.
© 2024 The Author(s). Journal of Cellular and Molecular Medicine published by Foundation for Cellular and Molecular Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

