SpliceTransformer predicts tissue-specific splicing linked to human diseases
- PMID: 39443442
- PMCID: PMC11500173
- DOI: 10.1038/s41467-024-53088-6
SpliceTransformer predicts tissue-specific splicing linked to human diseases
Abstract
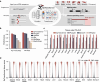
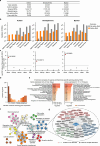
We present SpliceTransformer (SpTransformer), a deep-learning framework that predicts tissue-specific RNA splicing alterations linked to human diseases based on genomic sequence. SpTransformer outperforms all previous methods on splicing prediction. Application to approximately 1.3 million genetic variants in the ClinVar database reveals that splicing alterations account for 60% of intronic and synonymous pathogenic mutations, and occur at different frequencies across tissue types. Importantly, tissue-specific splicing alterations match their clinical manifestations independent of gene expression variation. We validate the enrichment in three brain disease datasets involving over 164,000 individuals. Additionally, we identify single nucleotide variations that cause brain-specific splicing alterations, and find disease-associated genes harboring these single nucleotide variations with distinct expression patterns involved in diverse biological processes. Finally, SpTransformer analysis of whole exon sequencing data from blood samples of patients with diabetic nephropathy predicts kidney-specific RNA splicing alterations with 83% accuracy, demonstrating the potential to infer disease-causing tissue-specific splicing events. SpTransformer provides a powerful tool to guide biological and clinical interpretations of human diseases.
© 2024. The Author(s).
Conflict of interest statement
The authors have submitted a patent application for the method. Other than this, the authors declare that they do not have any competing interests.
Figures




References
-
- Ahmed, M. S., Ikram, S., Bibi, N. & Mir, A. Hutchinson–Gilford progeria syndrome: a premature aging disease. Mol. Neurobiol.55, 4417–4427 (2018). - PubMed
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources

