Chemical inhibition of stomatal differentiation by perturbation of the master-regulatory bHLH heterodimer via an ACT-Like domain
- PMID: 39443460
- PMCID: PMC11500415
- DOI: 10.1038/s41467-024-53214-4
Chemical inhibition of stomatal differentiation by perturbation of the master-regulatory bHLH heterodimer via an ACT-Like domain
Abstract
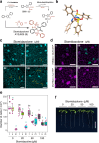
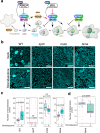
Selective perturbation of protein interactions with chemical compounds enables dissection and control of developmental processes. Differentiation of stomata, cellular valves vital for plant growth and survival, is specified by the basic-helix-loop-helix (bHLH) heterodimers. Harnessing a new amination reaction, we here report a synthesis, derivatization, target identification, and mode of action of an atypical doubly-sulfonylated imidazolone, Stomidazolone, which triggers stomatal stem cell arrest. Our forward chemical genetics followed by biophysical analyses elucidates that Stomidazolone directly binds to the C-terminal ACT-Like (ACTL) domain of MUTE, a master regulator of stomatal differentiation, and perturbs its heterodimerization with a partner bHLH, SCREAM in vitro and in plant cells. On the other hand, Stomidazolone analogs that are biologically inactive do not bind to MUTE or disrupt the SCREAM-MUTE heterodimers. Guided by structural docking modeling, we rationally design MUTE with reduced Stomidazolone binding. These engineered MUTE proteins are fully functional and confer Stomidazolone resistance in vivo. Our study identifies doubly-sulfonylated imidazolone as a direct inhibitor of the stomatal master regulator, further expanding the chemical space for perturbing bHLH-ACTL proteins to manipulate plant development.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Hetherington, A. M. & Woodward, F. I. The role of stomata in sensing and driving environmental change. Nature424, 901–908 (2003). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- JP16H01237/MEXT | Japan Society for the Promotion of Science (JSPS)
- JP17H06476/MEXT | Japan Society for the Promotion of Science (JSPS)
- JP19H00990/MEXT | Japan Society for the Promotion of Science (JSPS)
- Research Fellowship for Young Scientists DC1/MEXT | Japan Society for the Promotion of Science (JSPS)
- TORII/Howard Hughes Medical Institute (HHMI)
LinkOut - more resources
Full Text Sources

