LoDEI: a robust and sensitive tool to detect transcriptome-wide differential A-to-I editing in RNA-seq data
- PMID: 39443485
- PMCID: PMC11500352
- DOI: 10.1038/s41467-024-53298-y
LoDEI: a robust and sensitive tool to detect transcriptome-wide differential A-to-I editing in RNA-seq data
Abstract
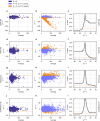
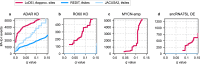
RNA editing is a highly conserved process. Adenosine deaminase acting on RNA (ADAR) mediated deamination of adenosine (A-to-I editing) is associated with human disease and immune checkpoint control. Functional implications of A-to-I editing are currently of broad interest to academic and industrial research as underscored by the fast-growing number of clinical studies applying base editors as therapeutic tools. Analyzing the dynamics of A-to-I editing, in a biological or therapeutic context, requires the sensitive detection of differential A-to-I editing, a currently unmet need. We introduce the local differential editing index (LoDEI) to detect differential A-to-I editing in RNA-seq datasets using a sliding-window approach coupled with an empirical q value calculation that detects more A-to-I editing sites at the same false-discovery rate compared to existing methods. LoDEI is validated on known and novel datasets revealing that the oncogene MYCN increases and that a specific small non-coding RNA reduces A-to-I editing.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

