Application of a metatranscriptomics technology, CSI-Dx, for the detection of pathogens associated with prosthetic joint infections
- PMID: 39443495
- PMCID: PMC11500344
- DOI: 10.1038/s41598-024-74375-8
Application of a metatranscriptomics technology, CSI-Dx, for the detection of pathogens associated with prosthetic joint infections
Abstract
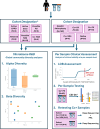
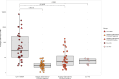
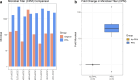
Preoperative identification of causal organism(s) is crucial for effective prosthetic joint infection treatment. Herein, we explore the clinical application of a novel metatranscriptomic (MT) workflow, CSI-Dx, to detect pathogens associated with prosthetic joint infection. MT provides insight into transcriptionally active microbes, overcoming limitations of culture-based and available molecular methods. This study included 340 human synovial fluid specimens subjected to CSI-Dx and traditional culture-based methods. Exploratory analyses were conducted to determine sensitivity and specificity of CSI-Dx for detecting clinically-relevant taxa. Our findings provide insights into the active microbial community composition of synovial fluid from arthroplasty patients and demonstrate the potential clinical utility of CSI-Dx for aiding prosthetic joint infection diagnosis. This approach offers potential for improved sensitivity and acceptable specificity compared to synovial fluid culture, enabling detection of culturable and non-culturable microorganisms. Furthermore, CSI-Dx provides valuable information on antimicrobial resistance gene expression. While further optimization is needed, integrating metatranscriptomic technologies like CSI-Dx into routine clinical practice can revolutionize prosthetic joint infection diagnosis by offering a comprehensive and active snapshot of associated pathogens.
© 2024. The Author(s).
Conflict of interest statement
The authors declare a conflict of interest. JRW, JCS, TTL, VT, JP, LP, SLCA, CYW, MH, AJS, and RL, are employees of Contamination Source Identification, LLC. KOT and SG have no relevant conflicts of interest.
Figures


References
MeSH terms
LinkOut - more resources
Full Text Sources

