Oral and gut microbiome profiles in people with early idiopathic Parkinson's disease
- PMID: 39443634
- PMCID: PMC11499922
- DOI: 10.1038/s43856-024-00630-8
Oral and gut microbiome profiles in people with early idiopathic Parkinson's disease
Abstract
Background: Early detection of Parkinson's disease (PD), a neurodegenerative disease with central and peripheral nerve involvement, ensures timely treatment access. Microbes influence nervous system health and are altered in PD.
Methods: We examined gut and mouth microbiomes from recently diagnosed patients in a geographically diverse, matched case-control, shotgun metagenomics study.
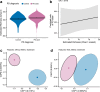
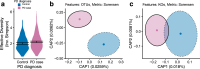
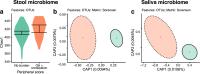
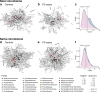
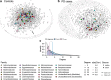
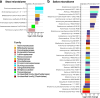
Results: Here, we show greater alpha-diversity in 445 PD patients versus 221 controls. The microbial signature of PD includes overabundance of 16 OTUs, including Streptococcus mutans and Bifidobacterium dentium, and depletion of 28 OTUs. Machine learning models indicate that subspecies level oral microbiome abundances best distinguish PD with reasonably high accuracy (area under the curve: 0.758). Microbial networks are disrupted in cases, with reduced connectivity between short-chain fatty acid-producing bacteria the the gut. Importantly, microbiome diversity metrics are associated with non-motor autonomic symptom severity.
Conclusions: Our results provide evidence that predictive oral PD microbiome signatures could possibly be used as biomarkers for the early detection of PD, particularly when there is peripheral nervous system involvement.
Plain language summary
Parkinson’s disease (PD) is a neurodegenerative disease that is characterized by both motor symptoms, such as tremors, and non-motor symptoms, such as constipation. Our aim was to determine whether there were differences in the number and types of microbes living in the saliva and intestines of people with and without PD. We saw significant differences in the microbial communities living in healthy controls compared to people with PD. Additionally, we found that the proportions of microbe types in saliva were the best at distinguishing between controls and cases, and identified the specific kinds of microbes that were driving this distinction. These results highlight the potential importance of the saliva microbiome in understanding the causes and symptomatology of PD.
© 2024. The Author(s).
Conflict of interest statement
The authors declare the following competing interests: K.S., M.J.K., M.W., S.A., T.F.S., P.F., J.T., M.V.H., and L.N.K. are employed by and hold stock or stock options in 23andMe, Inc.
Figures

References
LinkOut - more resources
Full Text Sources

