A Four-Gene Autophagy-Related Prognostic Model Signature and Its Association With Immune Phenotype in Lung Squamous Cell Carcinoma
- PMID: 39443755
- PMCID: PMC11499073
- DOI: 10.1002/cnr2.70000
A Four-Gene Autophagy-Related Prognostic Model Signature and Its Association With Immune Phenotype in Lung Squamous Cell Carcinoma
Abstract
Background: In the era of immunotherapy, there is a critical need for effective biomarkers to improve outcome prediction and guide treatment decisions for patients with lung squamous cell carcinoma (LUSC). We hypothesized that the immune contexture of LUSC may be influenced by tumor intrinsic events, such as autophagy.
Aims: We aimed to develop an autophagy-related risk signature and assess its predictive value for immune phenotype.
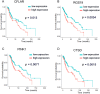
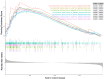
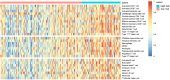
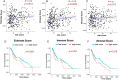
Methods and results: Expression profiles of autophagy-related genes (ARGs) in LUSC samples were obtained from the TCGA and GEO databases. Survival analyses were conducted to identify survival-related ARGs and construct a risk signature using the Random Forest algorithm. Four ARGs (CFLAR, RGS19, PINK1, and CTSD) with the most significant prognostic value were selected to construct the risk signature. Patients in the high-risk group exhibited worse prognosis than those in the low-risk group (p < 0.0001 in TCGA; p < 0.01 in GEO) and the risk score was identified as an independent prognostic factor. We observed that the high-risk group displayed an immune-suppressive status and showed higher levels of infiltrating regulatory T cells and macrophages, which are associated with poorer outcomes. Additionally, the risk score exhibited a significantly positive correlation with the expression of PD-1 and CTLA4, as well as the estimate score and immune score.
Conclusion: This study provided an effective autophagy-related prognostic signature, which could also predict the immune phenotype.
Keywords: autophagy; bioinformatic analysis; immune infiltration; immune landscape; lung squamous cell carcinoma; prognostic signature.
© 2024 The Author(s). Cancer Reports published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Siegel R. L., Miller K. D., and Jemal A., “Cancer Statistics, 2018,” CA: A Cancer Journal for Clinicians 68, no. 1 (2018): 7–30. - PubMed
-
- Torre L. A., Siegel R. L., and Jemal A., “Lung Cancer Statistics,” Advances in Experimental Medicine and Biology 893 (2016): 1–19. - PubMed
-
- Elanany M. M., Mostafa D., and Hamdy N. M., “Remodeled Tumor Immune Microenvironment (TIME) Parade via Natural Killer Cells Reprogramming in Breast Cancer,” Life Sciences 330 (2023): 121997. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

