The importance of family-based sampling for biobanks
- PMID: 39443775
- PMCID: PMC11623399
- DOI: 10.1038/s41586-024-07721-5
The importance of family-based sampling for biobanks
Abstract
Biobanks aim to improve our understanding of health and disease by collecting and analysing diverse biological and phenotypic information in large samples. So far, biobanks have largely pursued a population-based sampling strategy, where the individual is the unit of sampling, and familial relatedness occurs sporadically and by chance. This strategy has been remarkably efficient and successful, leading to thousands of scientific discoveries across multiple research domains, and plans for the next wave of biobanks are underway. In this Perspective, we discuss the strengths and limitations of a complementary sampling strategy for future biobanks based on oversampling of close genetic relatives. Such family-based samples facilitate research that clarifies causal relationships between putative risk factors and outcomes, particularly in estimates of genetic effects, because they enable analyses that reduce or eliminate confounding due to familial and demographic factors. Family-based biobank samples would also shed new light on fundamental questions across multiple fields that are often difficult to explore in population-based samples. Despite the potential for higher costs and greater analytical complexity, the many advantages of family-based samples should often outweigh their potential challenges.
© 2024. Springer Nature Limited.
Conflict of interest statement
Figures

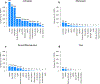
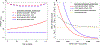
References
-
- Our Future Health. Our Future Health Study Protocol. https://s42615.pcdn.co/wp-content/uploads/Our-Future-Health-protocol-for... (2021).
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

