MdSVWC1, a new pattern recognition receptor triggers multiple defense mechanisms against invading bacteria in Musca domestica
- PMID: 39443921
- PMCID: PMC11515477
- DOI: 10.1186/s12915-024-02042-5
MdSVWC1, a new pattern recognition receptor triggers multiple defense mechanisms against invading bacteria in Musca domestica
Abstract
Background: Single-domain von Willebrand factor type C (SVWC) constitute a protein family predominantly identified in arthropods, characterized by a SVWC domain and involved in diverse physiological processes such as host defense, stress resistance, and nutrient metabolism. Nevertheless, the physiological mechanisms underlying these functions remain inadequately comprehended.
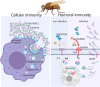
Results: A massive expansion of the SVWC gene family in Musca domestica (MdSVWC) was discovered, with a count of 35. MdSVWC1 was selected as the representative of the SVWC family for functional analysis, which led to the identification of the immune function of MdSVWC1 as a novel pattern recognition receptor. MdSVWC1 is highly expressed in both the fat body and intestines and displays acute induction upon bacterial infection. Recombinant MdSVWC1 binds to surfaces of both bacteria and yeast through the recognition of multiple pathogen-associated molecular patterns and exhibits Ca2+-dependent agglutination activity. MdSVWC1 mutant flies exhibited elevated mortality and hindered bacterial elimination following bacterial infection as a result of reduced hemocyte phagocytic capability and weakened expression of antimicrobial peptide (AMP) genes. In contrast, administration of recombinant MdSVWC1 provided protection to flies from bacterial challenges by promoting phagocytosis and AMP genes expression, thereby preventing bacterial colonization. MdSPN16, a serine protease inhibitor, was identified as a target protein of MdSVWC1. It was postulated that the interaction of MdSVWC1 with MdSPN16 would result in the activation of an extracellular proteolytic cascade, which would then initiate the Toll signaling pathway and facilitate the expression of AMP genes.
Conclusions: MdSVWC1 displays activity as a soluble pattern recognition receptor that regulates cellular and humoral immunity by recognizing microbial components and facilitating host defense.
Keywords: Musca domestica; Cellular immunity; Humoral immunity; Pattern recognition receptor; Single von Willebrand factor C-domain protein.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
A single von Willebrand factor C-domain protein acts as an extracellular pattern-recognition receptor in the river prawn Macrobrachium nipponense.J Biol Chem. 2020 Jul 24;295(30):10468-10477. doi: 10.1074/jbc.RA120.013270. Epub 2020 Jun 12. J Biol Chem. 2020. PMID: 32532819 Free PMC article.
-
Single von Willebrand factor C-domain protein-2 confers immune defense against bacterial infections in the silkworm, Bombyx mori.Int J Biol Macromol. 2024 Nov;279(Pt 2):135241. doi: 10.1016/j.ijbiomac.2024.135241. Epub 2024 Sep 2. Int J Biol Macromol. 2024. PMID: 39233173
-
Single von Willebrand factor C-domain protein confers host defense against white spot syndrome virus by functioning as a pattern recognition receptor in Macrobrachium nipponense.Int J Biol Macromol. 2023 Jun 30;241:124520. doi: 10.1016/j.ijbiomac.2023.124520. Epub 2023 Apr 19. Int J Biol Macromol. 2023. PMID: 37085073
-
Pattern recognition receptors from lepidopteran insects and their biological functions.Dev Comp Immunol. 2020 Jul;108:103688. doi: 10.1016/j.dci.2020.103688. Epub 2020 Mar 25. Dev Comp Immunol. 2020. PMID: 32222357 Review.
-
Single domain von Willebrand factor type C "cytokines" and the regulation of the stress/immune response in insects.Arch Insect Biochem Physiol. 2024 Jan;115(1):e22071. doi: 10.1002/arch.22071. Arch Insect Biochem Physiol. 2024. PMID: 38288483 Review.
Cited by
-
Resource reallocation under persistent immune activation drives trade-offs between life history and immunity in pirk-deficient Musca domestica.BMC Biol. 2025 Jul 22;23(1):220. doi: 10.1186/s12915-025-02324-6. BMC Biol. 2025. PMID: 40696362 Free PMC article.
References
-
- Nunes C, Sucena E, Koyama T. Endocrine regulation of immunity in insects. FEBS J. 2021;288:3928–47. - PubMed
-
- Hoffmann JA. The immune response of Drosophila. Nature. 2003;426(6962):33–8. - PubMed
-
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124(4):783–801. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

