Clustering-aided prediction of outcomes in patients with idiopathic pulmonary fibrosis
- PMID: 39443991
- PMCID: PMC11515489
- DOI: 10.1186/s12931-024-03015-6
Clustering-aided prediction of outcomes in patients with idiopathic pulmonary fibrosis
Abstract
Background: Blood biomarkers predictive of the progression of idiopathic pulmonary fibrosis (IPF) would be of value for research and clinical practice. We used data from the IPF-PRO Registry to investigate whether the addition of "omics" data to risk prediction models based on demographic and clinical characteristics improved prediction of the progression of IPF.
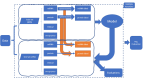
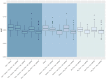
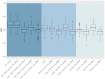
Methods: The IPF-PRO Registry enrolled patients with IPF at 46 sites across the US. Patients were followed prospectively. Median follow-up was 27.2 months. Prediction models for disease progression included omics data (proteins and microRNAs [miRNAs]), demographic factors and clinical factors, all assessed at enrollment. Data on proteins and miRNAs were included in the models either as raw values or based on clusters in various combinations. Least absolute shrinkage and selection operator (Lasso) Cox regression was applied for time-to-event composite outcomes and logistic regression with L1 penalty was applied for binary outcomes assessed at 1 year. Model performance was assessed using Harrell's C-index (for time-to-event outcomes) or area under the curve (for binary outcomes).
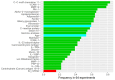
Results: Data were analyzed from 231 patients. The models based on demographic and clinical factors, with or without omics data, were the top-performing models for prediction of all the time-to-event outcomes. Relative changes in average C-index after incorporating omics data into models based on demographic and clinical factors ranged from 1.7 to 3.2%. Of the blood biomarkers, surfactant protein-D, serine protease inhibitor A7 and matrix metalloproteinase-9 (MMP-9) were among the top predictors of the outcomes. For the binary outcomes, models based on demographics alone and models based on demographics plus omics data had similar performances. Of the blood biomarkers, CC motif chemokine 11, vascular cell adhesion protein-1, adiponectin, carcinoembryonic antigen and MMP-9 were the most important predictors of the binary outcomes.
Conclusions: We identified circulating protein and miRNA biomarkers associated with the progression of IPF. However, the integration of omics data into prediction models that included demographic and clinical factors did not materially improve the performance of the models.
Trial registration: ClinicalTrials.gov; No: NCT01915511; registered August 5, 2013; URL: www.
Clinicaltrials: gov .
Keywords: Biomarkers; Disease progression; Interstitial lung disease.
© 2024. The Author(s).
Conflict of interest statement
Lijun Wang, Peitao Wu, Yi Liu, and Divya C Patel are employees of Boehringer Ingelheim Pharmaceuticals, Inc. Thomas B Leonard was an employee of Boehringer Ingelheim Pharmaceuticals, Inc at the time that these analyses were planned and conducted. Hongyu Zhao has no competing interests other than the funding of this project by Boehringer Ingelheim Pharmaceuticals, Inc.
Figures


References
-
- Fainberg HP, Oldham JM, Molyneau PL, Allen RJ, Kraven LM, Fahy WA, et al. Forced vital capacity trajectories in patients with idiopathic pulmonary fibrosis: a secondary analysis of a multicentre, prospective, observational cohort. Lancet Digit Health. 2022;4:e862–72. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

