Comparative Analysis of Transcription Factors TWIST2, GATA3, and HES5 in Glioblastoma Multiforme : Evaluating Biomarker Potential and Therapeutic Targets Using in Silico Methods
- PMID: 39444320
- PMCID: PMC11924635
- DOI: 10.3340/jkns.2024.0149
Comparative Analysis of Transcription Factors TWIST2, GATA3, and HES5 in Glioblastoma Multiforme : Evaluating Biomarker Potential and Therapeutic Targets Using in Silico Methods
Abstract
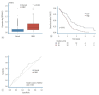
Objective: Glioblastoma multiforme (GBM) is characterized by substantial heterogeneity and limited therapeutic options. As molecular approaches to central nervous system tumors have gained prominence, this study examined the roles of three genes, TWIST2, GATA3, and HES5, known to be involved in oncogenesis, developmental processes, and maintenance of cancer stem cell properties, which have not yet been extensively studied in GBM. This study is the first to present gene expression data for TWIST2, GATA3, and HES5 specifically within the context of GBM patient survival.
Methods: Gene expression data for TWIST2, GATA3, and HES5 were collected from GBM and normal brain tissues using datasets from The Cancer Genome Atlas via the Genomic Data Commons portal and the Genotype-Tissue Expression database. These data were rigorously analyzed using in silico methods.
Results: All three genes were significantly more expressed in GBM tissues than in normal tissues. TWIST2 and GATA3 were linked to lower survival rates in GBM patients. Interestingly, higher HES5 levels were associated with better survival rates, suggesting a complex role that needs more investigation.
Conclusion: This study shows that TWIST2, GATA3, and HES5 could help predict outcomes in GBM patients. Our multigene model offers a better understanding of GBM and points to new treatment options, bringing hope for improved therapies and patient outcomes. This research advances our knowledge of GBM and highlights the potential of molecular diagnostics in oncology.
Keywords: GATA3; Gene expression; Glioblastoma; HES5; TWIST2.
Conflict of interest statement
No potential conflict of interest relevant to this article was reported.
Figures



Similar articles
-
Analysis of the cancer genome atlas (TCGA) database identifies an inverse relationship between interleukin-13 receptor α1 and α2 gene expression and poor prognosis and drug resistance in subjects with glioblastoma multiforme.J Neurooncol. 2018 Feb;136(3):463-474. doi: 10.1007/s11060-017-2680-9. Epub 2017 Nov 22. J Neurooncol. 2018. PMID: 29168083 Free PMC article.
-
A Multistep In Silico Approach Identifies Potential Glioblastoma Drug Candidates via Inclusive Molecular Targeting of Glioblastoma Stem Cells.Mol Neurobiol. 2024 Nov;61(11):9253-9271. doi: 10.1007/s12035-024-04139-y. Epub 2024 Apr 15. Mol Neurobiol. 2024. PMID: 38619743
-
SLC12A5 as a novel potential biomarker of glioblastoma multiforme.Mol Biol Rep. 2023 May;50(5):4285-4299. doi: 10.1007/s11033-023-08371-y. Epub 2023 Mar 14. Mol Biol Rep. 2023. PMID: 36917367
-
Current Perspectives on Therapies, Including Drug Delivery Systems, for Managing Glioblastoma Multiforme.ACS Chem Neurosci. 2020 Oct 7;11(19):2962-2977. doi: 10.1021/acschemneuro.0c00555. Epub 2020 Sep 18. ACS Chem Neurosci. 2020. PMID: 32945654 Review.
-
An update on the molecular biology of glioblastoma, with clinical implications and progress in its treatment.Cancer Commun (Lond). 2022 Nov;42(11):1083-1111. doi: 10.1002/cac2.12361. Epub 2022 Sep 21. Cancer Commun (Lond). 2022. PMID: 36129048 Free PMC article. Review.
References
-
- Basak O, Taylor V. Identification of self-replicating multipotent progenitors in the embryonic nervous system by high Notch activity and Hes5 expression. Eur J Neurosci. 2007;25:1006–1022. - PubMed
-
- Debacker C, Catala M, Labastie MC. Embryonic expression of the human GATA-3 gene. Mech Dev. 1999;85:183–187. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

