To Develop Biomarkers for Diabetic Nephropathy Based on Genes Related to Fibrosis and Propionate Metabolism and Their Functional Validation
- PMID: 39444490
- PMCID: PMC11498995
- DOI: 10.1155/2024/9066326
To Develop Biomarkers for Diabetic Nephropathy Based on Genes Related to Fibrosis and Propionate Metabolism and Their Functional Validation
Abstract
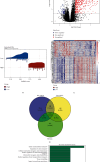
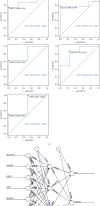
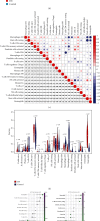
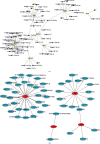
Propionate metabolism is important in the development of diabetes, and fibrosis plays an important role in diabetic nephropathy (DN). However, there are no studies on biomarkers related to fibrosis and propionate metabolism in DN. Hence, the current research is aimed at evaluating biomarkers associated with fibrosis and propionate metabolism and to explore their effect on DN progression. The GSE96804 (DN : control = 41 : 20) and GSE104948 (DN : control = 7 : 18) DN-related datasets and 924 propionate metabolism-related genes (PMRGs) and 656 fibrosis-related genes (FRGs) were acquired from the public database. First, DN differentially expressed genes (DN-DEGs) between the DN and control samples were sifted out via differential expression analysis. The PMRG scores of the DN samples were calculated based on PMRGs. The samples were divided into the high and low PMRG score groups according to the median scores. The PM-DEGs between the two groups were screened out. Second, the intersection of DN-DEGs, PM-DEGs, and FRGs was taken to yield intersected genes. Random forest (RF) and recursive feature elimination (RFE) analyses of the intersected genes were performed to sift out biomarkers. Then, single gene set enrichment analysis was conducted. Finally, immunoinfiltrative analysis was performed, and the transcription factor (TF)-microRNA (miRNA)-mRNA regulatory network and the drug-gene interaction network were constructed. There were 2633 DN-DEGs between the DN and control samples and 515 PM-DEGs between the high and low PMRG score groups. In total, 10 intersected genes were gained after taking the intersection of DN-DEGs, PM-DEGs, and FRGs. Seven biomarkers, namely, SLC37A4, ACOX2, GPD1, angiotensin-converting enzyme 2 (ACE2), SLC9A3, AGT, and PLG, were acquired via RF and RFE analyses, and they were found to be involved in various mechanisms such as glomerulus development, fatty acid metabolism, and peroxisome. The seven biomarkers were positively correlated with neutrophils. Moreover, 8 TFs, 60 miRNAs, and 7 mRNAs formed the TF-miRNA-mRNA regulatory network, including USF1-hsa-mir-1296-5p-AGT and HIF1A-hsa-mir-449a-5p-ACE2. The drug-gene network contained UROKINASE-PLG, ATENOLOL-AGT, and other interaction relationship pairs. Via bioinformatic analyses, the risk of fibrosis and propionate metabolism-related biomarkers in DN were explored, thereby providing novel ideas for research related to DN diagnosis and treatment.
Copyright © 2024 Sha Li et al.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

