Asymmetric Monoreduction of α,β-Dicarbonyls to α-Hydroxy Carbonyls by Ene Reductases
- PMID: 39444529
- PMCID: PMC11494505
- DOI: 10.1021/acscatal.4c04676
Asymmetric Monoreduction of α,β-Dicarbonyls to α-Hydroxy Carbonyls by Ene Reductases
Abstract
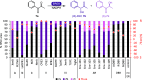
Ene reductases (EREDs) catalyze asymmetric reduction with exquisite chemo-, stereo-, and regioselectivity. Recent discoveries led to unlocking other types of reactivities toward oxime reduction and reductive C-C bond formation. Exploring nontypical reactions can further expand the biocatalytic knowledgebase, and evidence alludes to yet another variant reaction where flavin mononucleotide (FMN)-bound ERs from the old yellow enzyme family (OYE) have unconventional activity with α,β-dicarbonyl substrates. In this study, we demonstrate the nonconventional stereoselective monoreduction of α,β-dicarbonyl to the corresponding chiral hydroxycarbonyl, which are valuable building blocks for asymmetric synthesis. We explored ten α,β-dicarbonyl aliphatic, cyclic, or aromatic compounds and tested their reduction with five OYEs and one nonflavin-dependent double bond reductase (DBR). Only GluER reduced aliphatic α,β-dicarbonyls, with up to 19% conversion of 2,3-hexanedione to 2-hydroxyhexan-3-one with an R-selectivity of 83% ee. The best substrate was the aromatic α,β-dicarbonyl 1-phenyl-1,2-propanedione, with 91% conversion to phenylacetylcarbinol using OYE3 with R-selectivity >99.9% ee. Michaelis-Menten kinetics for 1-phenyl-1,2-propanedione with OYE3 gave a turnover k cat of 0.71 ± 0.03 s-1 and a K m of 2.46 ± 0.25 mM. Twenty-four EREDs from multiple classes of OYEs and DBRs were further screened on 1-phenyl-1,2-propanedione, showing that class II OYEs (OYE3-like) have the best overall selectivity and conversion. EPR studies detected no radical signal, whereas NMR studies with deuterium labeling indicate proton incorporation at the benzylic carbonyl carbon from the solvent and not the FMN hydride. A crystal structure of OYE2 with 1.5 Å resolution was obtained, and docking studies showed a productive pose with the substrate.
© 2024 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures





References
-
- Warburg O.; Christian W. On a New Oxidation Enzyme. Naturwissenschaften 1932, 20, 980–981. 10.1007/BF01504728. - DOI
-
- Bender S. G.; Hyster T. K. Pyridylmethyl Radicals for Enantioselective Alkene Hydroalkylation Using “Ene”-Reductases. ACS Catal. 2023, 13, 14680–14684. 10.1021/acscatal.3c03771. - DOI
-
- Böhmer S.; Marx C.; Gómez-Baraibar Á.; Nowaczyk M. M.; Tischler D.; Hemschemeier A.; Happe T. Evolutionary Diverse Chlamydomonas Reinhardtii Old Yellow Enzymes Reveal Distinctive Catalytic Properties and Potential for Whole-Cell Biotransformations. Algal Res. 2020, 50, 101970. 10.1016/j.algal.2020.101970. - DOI
-
- Schittmayer M.; Glieder A.; Uhl M. K.; Winkler A.; Zach S.; Schrittwieser J. H.; Kroutil W.; MacHeroux P.; Gruber K.; Kambourakis S.; Rozzell J. D.; Winkler M. Old Yellow Enzyme-Catalyzed Dehydrogenation of Saturated Ketones. Adv. Synth. Catal. 2011, 353, 268–274. 10.1002/adsc.201000862. - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous
