Genetic diversity and evolution of porcine hemagglutinating encephalomyelitis virus in Guangxi province of China during 2021-2024
- PMID: 39444682
- PMCID: PMC11496168
- DOI: 10.3389/fmicb.2024.1474552
Genetic diversity and evolution of porcine hemagglutinating encephalomyelitis virus in Guangxi province of China during 2021-2024
Abstract
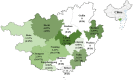
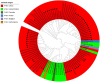
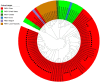
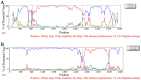
Porcine hemagglutinating encephalomyelitis virus (PHEV) is the only known porcine neurotropic coronavirus, which is prevalent worldwide at present. It is of great significance to understand the genetic and evolutionary characteristics of PHEV in order to perform effective measures for prevention and control of this disease. In this study, a total of 6,986 tissue samples and nasopharyngeal swabs were collected from different regions of Guangxi province in southern China during 2021-2024, and were tested for PHEV using a quadruplex RT-qPCR. The positivity rate of PHEV was 2.81% (196/6,986), of which tissue samples and nasopharyngeal swabs had 2.05% (87/4,246) and 3.98% (109/2,740) positivity rates, respectively. Fifty PHEV positive samples were selected for PCR amplification and gene sequencing. Sequence analysis revealed that the nucleotide homology and amino acid similarities of S, M, and N genes were 94.3%-99.3% and 92.3%-99.2%, 95.0%-99.7% and 94.7%-100.0%, 94.0%-99.5% and 93.5%-99.3%, respectively, indicating M and N genes were more conservative than S gene. Phylogenetic trees based on these three genes revealed that PHEV strains from different countries could be divided into two groups G1 and G2, and the PHEV strains from Guangxi province obtained in this study distributed in subgroups G1c and G2b. Bayesian analysis revealed that the population size of PHEV has been in a relatively stable state since its discovery until it expanded sharply around 2015, and still on the slow rise thereafter. S gene sequences analysis indicated that PHEV strains existed variation of mutation, and recombination. The results indicated that the prevalent PHEV strains in Guangxi province had complex evolutionary trajectories and high genetic diversity. To the best of our knowledge, this is the first report on the genetic and evolutionary characteristics of PHEV in southern China.
Keywords: coronavirus; genetic evolution; phylogenetic analysis; porcine hemagglutinating encephalomyelitis virus; recombination.
Copyright © 2024 Shi, Hu, Long, Shi, Pan, Feng, Li and Yin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Alsop J. E. (2006). A presumptive case of vomiting and wasting disease in a swine nucleus herd. J. Swine Health Product. 14, 97–100. doi: 10.54846/jshap/459 - DOI
-
- Bahoussi A. N., Guo Y. Y., Shi R. Z., Wang P. H., Li Y. Q., Wu C. X., et al. (2022). Genetic characteristics of porcine hemagglutinating encephalomyelitis coronavirus: identification of naturally occurring mutations between 1970 and 2015. Front. Microbiol. 13:860851. doi: 10.3389/fmicb.2022.860851, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

