Induced degradation of SNAP-fusion proteins
- PMID: 39444693
- PMCID: PMC11494418
- DOI: 10.1039/d4cb00184b
Induced degradation of SNAP-fusion proteins
Abstract
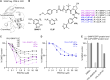
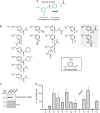
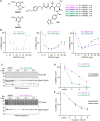
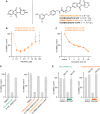
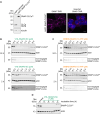
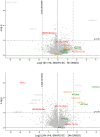
Self-labeling protein tags are an efficient means to visualize, manipulate, and isolate engineered fusion proteins with suitable chemical probes. The SNAP-tag, which covalently conjugates to benzyl-guanine and -chloropyrimidine derivatives is used extensively in fluorescence microscopy, given the availability of suitable SNAP-ligand-based probes. Here, we extend the applicability of the SNAP-tag to targeted protein degradation. We developed a set of SNAP PROteolysis TArgeting Chimeras (SNAP-PROTACs), which recruit the VHL or CRBN-ubiquitin E3 ligases to induce the degradation of SNAP-fusion proteins. Endogenous tagging enabled the visualization and the selective depletion of a SNAP-clathrin light chain fusion protein using SNAP-PROTACs. The addition of PROTACs to the SNAP-tag reagent toolbox facilitates the comprehensive analysis of protein function with a single gene tagging event.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts of interest to declare.
Figures
References
LinkOut - more resources
Full Text Sources

