Identification of key proteins in early-onset Alzheimer's disease based on WGCNA
- PMID: 39444808
- PMCID: PMC11496171
- DOI: 10.3389/fnagi.2024.1412222
Identification of key proteins in early-onset Alzheimer's disease based on WGCNA
Abstract
Introduction: Early-onset Alzheimer's disease (EOAD) is sporadic, highly heterogeneous, and its underlying pathogenic mechanisms remain largely elusive. Proteomics research aims to uncover the biological processes and key proteins involved in disease progression. However, no proteomic studies to date have specifically focused on EOAD brain tissue.
Method: We integrated proteomic data from brain tissues of two Alzheimer's disease (AD) cohorts and constructed a protein co-expression network using weighted gene co-expression network analysis (WGCNA). We identified modules associated with EOAD, conducted functional enrichment analysis to understand the biological processes involved in EOAD, and pinpointed potential key proteins within the core modules most closely linked to AD pathology.
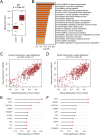
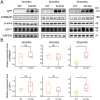
Results: In this study, we identified a total of 2,749 proteins associated with EOAD. Through protein co-expression network analysis, we discovered 41 distinct co-expression modules. Notably, the proteins within the core module most closely linked to AD pathology were significantly enriched in neutrophil degranulation. Additionally, we identified two potential key proteins within this core module that have not been previously reported in AD and validated their expression levels in 5xFAD mice.
Conclusion: In summary, through a protein co-expression network analysis, we identified EOAD-related biological processes and molecular pathways, and screened and validated two key proteins, ERBB2IP and LSP1. These proteins may play an important role in the progression of EOAD, suggesting they could serve as potential therapeutic targets for the disease.
Keywords: WGCNA; biological process; early-onset Alzheimer disease; proteome; therapeutic target.
Copyright © 2024 Li, Wang, Wang and Tang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Miscellaneous

