Prognostic model of lung adenocarcinoma based on immunoprognosis-related genes and related drug prediction
- PMID: 39444861
- PMCID: PMC11494575
- DOI: 10.21037/jtd-24-569
Prognostic model of lung adenocarcinoma based on immunoprognosis-related genes and related drug prediction
Abstract
Background: Lung cancer (LC) is the most common malignant tumor in the world, and lung adenocarcinoma (LUAD) is the most common type of LC. Immune microenvironment plays a critical role in cancer from onset to relapse. We aimed to identify an effective immune-related prediction model for assessing prognosis and predicting the relevant tumor therapeutic drugs.
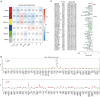
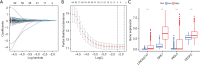
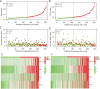
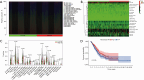
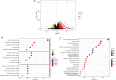
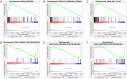
Methods: According to the RNA sequencing data of LUAD transcriptome in The Cancer Genome Atlas (TCGA) database and the immune-related genes obtained from IMMPORT (The Immunology Database and Analysis Portal) database, immune prognosis-related genes were screened. Weighted gene co-expression network analysis (WGCNA) identified hub genes in differentially expressed immune-related genes (DEIRGs). Least absolute shrinkage and selection operator (LASSO) Cox and ten rounds of cross-validation were used to screen core genes to establish a prognostic model, and in situ hybridization was used to verify the expression of core genes in LUAD. Then the patients from the TCGA database were divided into high-risk group and low-risk group. The survival, tumor microenvironment (TME) and immune cell infiltration of different groups were further analyzed, and the differential genes between the two groups were analyzed by gene ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Set Enrichment Analysis (GSEA) enrichment analyses. Finally, the small molecular drugs that can inhibit the prognosis of LUAD were screened by Connectivity Map (CMAP), and the therapeutic mechanism of small molecular drug oxibendazole was verified by Cell Counting Kit-8 (CCK-8) experiment.
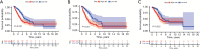
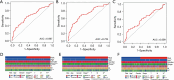
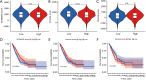
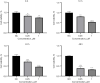
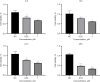
Results: A four-immunoprognosis-related gene model was established to forecast the overall survival (OS) of LUAD through LASSO Cox regression and ten rounds of cross-validation analysis. This prognostic model stratified LUAD patients into low-risk and high-risk groups. According to the findings of the survival analysis, the low-risk group had a greater OS than the high-risk group and the content of immune cells in LUAD was corrected with the survival prognosis of patients. Univariate and multivariate Cox regression also revealed that the prognostic model was an independent prognosis factor in LUAD. Five kinds of small molecular drugs which can inhibit the prognosis of LUAD were screened by CMAP. As shown by CCK-8 test, the small molecular drug "oxibendazole" can effectively inhibit the proliferation of LUAD cells.
Conclusions: Based on immune-related prognostic genes, a new prognostic model for LUAD was constructed. Oxibendazole can inhibit the proliferation of LUAD cells, which provides a new idea for the treatment of LUAD.
Keywords: Oxibendazole; The Cancer Genome Atlas (TCGA); immunoprognostic genes; lung adenocarcinoma (LUAD); prognostic model.
2024 AME Publishing Company. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-24-569/coif). Y.J. reports that this project was supported by National Natural Science Foundation of China, Youth Science Foundation Project (No. 82300412). The other authors have no conflicts of interest to declare.
Figures


References
LinkOut - more resources
Full Text Sources
Research Materials
