Bacterial Analysis of the Whole Blood in Chinese Healthy Donors Using 16S rDNA-Targeted Metagenomic Sequencing
- PMID: 39444936
- PMCID: PMC11498981
- DOI: 10.1155/2024/6635560
Bacterial Analysis of the Whole Blood in Chinese Healthy Donors Using 16S rDNA-Targeted Metagenomic Sequencing
Abstract
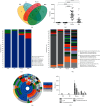
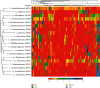
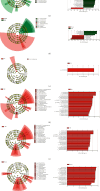
Background: The presence of bacteria in the blood of healthy individuals remains controversial. This study explored the comprehensive bacterial profiles and specific biomarkers in different components of healthy Chinese blood donors. Methods: A total of 5230 whole blood (WB) specimens were collected. Among them, 5200 random samples were pooled into 26 mixed samples for bacterial profile analysis. The remaining 30 random samples were divided into 4 groups based on components: WB, plasma, red blood cells (RBCs), and buffy coat (BC). Subsequently, the amplicons of the bacterial 16S rDNA V3-V4 fragments were sequenced to measure the diversity and composition of the bacteria using next-generation sequencing. Results: The bacterial DNAs in the blood primarily originated from the Proteobacteria phylum. A total of 301 species of bacterial DNA were found in blood specimens, with 46 species being present among all groups. A significantly higher abundance of bacterial DNA was found in the plasma and RBCs compared to those in BC and WB. However, the plasma and RBC groups showed significantly higher species diversity and richness compared to the BC and WB groups. In addition, the WB group had a significantly different community structure and composition compared to the plasma and RBC groups but was similar to the BC group. Conclusion: The presence of bacterial DNA fragments was confirmed in blood from healthy Chinese donors. The bacterial DNA fragments enriched in plasma showed the highest diversity, followed by RBC, WB, and BC. These results provide a foundation for further research on the microbiome in the blood of healthy individuals.
Keywords: 16S rDNA; Chinese blood donors; blood components; blood microbiome; whole blood.
Copyright © 2024 Jingjing Zhang et al.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
LinkOut - more resources
Full Text Sources

