Prognostic modeling of hepatocellular carcinoma based on T-cell proliferation regulators: a bioinformatics approach
- PMID: 39445019
- PMCID: PMC11496079
- DOI: 10.3389/fimmu.2024.1444091
Prognostic modeling of hepatocellular carcinoma based on T-cell proliferation regulators: a bioinformatics approach
Abstract
Background: The prognostic value and immune significance of T-cell proliferation regulators (TCRs) in hepatocellular carcinoma (HCC) have not been previously reported. This study aimed to develop a new prognostic model based on TCRs in patients with HCC.
Method: This study used The Cancer Genome Atlas-Liver Hepatocellular Carcinoma (TCGA-LIHC) and International Cancer Genome Consortium-Liver Cancer-Riken, Japan (ICGC-LIRI-JP) datasets along with TCRs. Differentially expressed TCRs (DE-TCRs) were identified by intersecting TCRs and differentially expressed genes between HCC and non-cancerous samples. Prognostic genes were determined using Cox regression analysis and were used to construct a risk model for HCC. Kaplan-Meier survival analysis was performed to assess the difference in survival between high-risk and low-risk groups. Receiver operating characteristic curve was used to assess the validity of risk model, as well as for testing in the ICGC-LIRI-JP dataset. Additionally, independent prognostic factors were identified using multivariate Cox regression analysis and proportional hazards assumption, and they were used to construct a nomogram model. TCGA-LIHC dataset was subjected to tumor microenvironment analysis, drug sensitivity analysis, gene set variation analysis, and immune correlation analysis. The prognostic genes were analyzed using consensus clustering analysis, mutation analysis, copy number variation analysis, gene set enrichment analysis, and molecular prediction analysis.
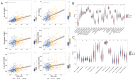
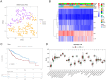
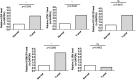
Results: Among the 18 DE-TCRs, six genes (DCLRE1B, RAN, HOMER1, ADA, CDK1, and IL1RN) could predict the prognosis of HCC. A risk model that can accurately predict HCC prognosis was established based on these genes. An efficient nomogram model was also developed using clinical traits and risk scores. Immune-related analyses revealed that 39 immune checkpoints exhibited differential expression between the high-risk and low-risk groups. The rate of immunotherapy response was low in patients belonging to the high-risk group. Patients with HCC were further divided into cluster 1 and cluster 2 based on prognostic genes. Mutation analysis revealed that HOMER1 and CDK1 harbored missense mutations. DCLRE1B exhibited an increased copy number, whereas RAN exhibited a decreased copy number. The prognostic genes were significantly enriched in tryptophan metabolism pathways.
Conclusions: This bioinformatics analysis identified six TCR genes associated with HCC prognosis that can serve as diagnostic markers and therapeutic targets for HCC.
Keywords: GEO; T-cell proliferation regulators; bioinformatic; hepatocellular carcinoma; prognostic model.
Copyright © 2024 Hai, Bai, Luo, Liu, Ma, Ma and Ding.
Conflict of interest statement
ZM is employed by Weiluo Microbial Pathogens Monitoring Technology Co., Ltd. of Beijing. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

